Abstract
Purpose
Genomic alterations are fundamental for molecular-guided therapy in patients with breast and lung cancer. However, the turn-around time of standard next-generation sequencing assays is a limiting factor in the timely delivery of genomic information for clinical decision-making.
Methods
In this study, we evaluated genomic alterations in 54 cerebrospinal fluid samples from 33 patients with metastatic lung cancer and metastatic breast cancer to the brain using the Oncomine Precision Assay on the Genexus sequencer. There were nine patients with samples collected at multiple time points.
Results
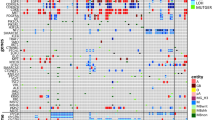
Cell-free total nucleic acids (cfTNA) were extracted from CSF (0.1–11.2 ng/μl). Median base coverage was 31,963× with cfDNA input ranging from 2 to 20 ng. Mutations were detected in 30/54 CSF samples. Nineteen (19/24) samples with no mutations detected had suboptimal DNA input (< 20 ng). The EGFR exon-19 deletion and PIK3CA mutations were detected in two patients with increasing mutant allele fraction over time, highlighting the potential of CSF-cfTNA analysis for monitoring patients. Moreover, the EGFR T790M mutation was detected in one patient with prior EGFR inhibitor treatment. Additionally, ESR1 D538G and ESR1::CCDC170 alterations, associated with endocrine therapy resistance, were detected in 2 mBC patients. The average TAT from cfTNA-to-results was < 24 h.
Conclusion
In summary, our results indicate that CSF-cfTNA analysis with the Genexus-OPA can provide clinically relevant information in patients with brain metastases with short TAT.




Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article.
References
Barnholtz-Sloan JS et al (2004) Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J Clin Oncol 22:2865–2872
Schouten LJ et al (2002) Incidence of brain metastases in a cohort of patients with carcinoma of the breast, colon, kidney, and lung and melanoma. Cancer 94:2698–2705
Ali S et al (2021) Molecular profiles of brain metastases: a focus on heterogeneity. Cancers 13:2645
Heitzer E et al (2019) Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 20:71–88
De Mattos-Arruda L et al (2015) Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 6:1–6
Pan C et al (2019) Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol 137:297–306
Wang Y et al (2015) Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci USA 112:9704–9709
Bettegowda C et al (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6(224):24
Pan W et al (2015) Brain tumor mutations detected in cerebral spinal fluid. Clin Chem 61:514–522
Takayasu T et al (2020) Cerebrospinal fluid ctDNA and metabolites are informative biomarkers for the evaluation of CNS germ cell tumors. Sci Rep 10:14326
Zorofchian S et al (2018) Detection of the MYD88p.L265P mutation in the CSF of a patient with secondary central nervous system lymphoma. Front Oncol 8:1–6
Fujita Y et al (2022) IDH1 p. R132H ctDNA and D-2-hydroxyglutarate as CSF biomarkers in patients with IDH-mutant gliomas. J Neurooncol 159:261–270
Pentsova EI et al (2016) Evaluating cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol 34:2404–2415
Ballester LY et al (2018) Evaluating circulating tumor DNA from the cerebrospinal fluid of patients with melanoma and leptomeningeal disease. J Neuropathol Exp Neurol 77:628–635
Shah M et al (2021) Evaluation of the Oncomine pan-cancer cell-free assay for analyzing circulating tumor DNA in the cerebrospinal fluid in patients with central nervous system malignancies. J Mol Diagn 23:171–180
Kou T et al (2017) Clinical sequencing using a next-generation sequencing-based multiplex gene assay in patients with advanced solid tumors. Cancer Sci 108:1440–1446
Thomson AH et al (2016) Changing molecular profile of brain metastases compared with matched breast primary cancers and impact on clinical outcomes. Br J Cancer 114:793–800
Ferguson SD et al (2018) Profiles of brain metastases: prioritization of therapeutic targets. Int J Cancer 143:3019–3026
Wu J et al (2022) Cerebrospinal fluid circulating tumor DNA depicts profiling of brain metastasis in NSCLC. Mol Oncol. https://doi.org/10.1002/1878-0261.13357
Ma C et al (2020) Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Thorac Cancer 11:588–593
Li L et al (2020) Therapeutic role of recurrent ESR1-CCDC170 gene fusions in breast cancer endocrine resistance. Breast Cancer Res 22:84
Funding
Research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health under award number K08CA241651 and this work was partly supported by the generous philanthropic contributions to The University of Texas MD Anderson Cancer Center Glioblastoma Moon Shots Program™. This work was partially supported by an Oncomine Clinical Research Grant from Thermo Fisher Scientific (awarded to LYB).
Author information
Authors and Affiliations
Contributions
JJZ, SRC, FL and YE—contributed to sample collection. AD, YE, JJZ—patient consent. MS and AD—performed sample processing and cfDNA extraction. SA—performed NGS experiments and prepared initial manuscript draft. PP and LN—performed sample annotation and collection of clinical information. DW-contributed to the preparation of figures and data analysis. LB, RL, DD and YE—Project conception, experimental design and manuscript writing. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose. The authors have no competing interests to declare that are relevant to the content of this article. All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript. The authors have no financial or proprietary interests in any material discussed in this article.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the local ethics regulations and approvals and with the 1964 Helsinki declaration and its later amendments. All procedures were performed in accordance with local institutional review board (IRB MDACC and UTHealth) guidelines.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11060_2023_4487_MOESM1_ESM.jpg
Supplementary fig 1: The overall survival of patients with positive and negative CSF-ctDNA results. (A) Survival analysis of patients with mLC. There were 10 patients with CSF ctDNA positive and 11 patients with CSF ctDNA negative results. The median survival was 37 months and 45 months for CSF ctDNA positive and CSF ctDNA negative patients, respectively. (B) Survival analysis of patients with mBC. There were 9 patients with CSF ctDNA positive and 3 with CSF ctDNA negative results. The median survival was undefined for both CSF ctDNA positive and CSF ctDNA negative mBC patients. Supplementary file1 (JPG 217 kb)
11060_2023_4487_MOESM2_ESM.jpg
Supplementary fig 2A–E: MRI results and CSF ctDNA results for patients with multiple CSF samples. (A) MRI results and CSF ctDNA results from mLC sample #1, (B) MRI results and CSF ctDNA results from mLC sample #3, (C) MRI results and CSF ctDNA results from mBC sample #2, (D) MRI results and CSF ctDNA results from mBC sample #5, (E) MRI results and CSF ctDNA results from mBC sample #6. Supplementary file2 (JPG 1129 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Arjuna, S., Shah, M., Dono, A. et al. Rapid detection of mutations in CSF-cfTNA with the Genexus Integrated Sequencer. J Neurooncol 166, 39–49 (2024). https://doi.org/10.1007/s11060-023-04487-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-023-04487-0