Abstract
Purpose
The primary purpose of this study was to utilize machine learning (ML) models to create a web application that can predict survival outcomes for patients diagnosed with atypical and anaplastic meningiomas.
Methods
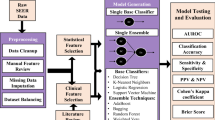
In this retrospective cohort study, patients diagnosed with WHO grade II and III meningiomas were selected from the National Cancer Database (NCDB) to analyze survival outcomes at 12, 36, and 60 months. Five machine learning algorithms - TabPFN, TabNet, XGBoost, LightGBM, and Random Forest were employed and optimized using the Optuna library for hyperparameter tuning. The top-performing models were then deployed into our web-based application.
Results
From the NCDB, 12,197 adult patients diagnosed with histologically confirmed WHO grade II and III meningiomas were retrieved. The mean age was 61 (± 20), and 6,847 (56.1%) of these were females. Performance evaluation indicated that the top-performing models for each outcome were the models built with the TabPFN algorithm. The TabPFN models yielded area under the receiver operating characteristic (AUROC) values of 0.805, 0.781, and 0.815 in predicting 12-, 36-, and 60-month mortality, respectively.
Conclusion
With the continuous growth of neuro-oncology data, ML algorithms act as key tools in predicting survival outcomes for WHO grade II and III meningioma patients. By incorporating these interpretable models into a web application, we can practically utilize them to improve risk evaluation and prognosis for meningioma patients.



Similar content being viewed by others
Data Availability
Restrictions apply to the availability of these data. Data were obtained from the NCDB, a prospectively maintained repository collaboratively developed by the Commission on Cancer (CoC) of the American College of Surgeons and the American Cancer Society.
References
Ostrom QT, Cioffi G, Waite K et al (2021) CBTRUS Statistical Report: primary brain and other Central Nervous System Tumors diagnosed in the United States in 2014–2018. Neuro-Oncol 23:iii1–iii105. https://doi.org/10.1093/neuonc/noab200
Baldi I, Engelhardt J, Bonnet C et al (2018) Epidemiology of meningiomas. Neurochirurgie 64:5–14. https://doi.org/10.1016/j.neuchi.2014.05.006
Lin D, Lin J, Deng X et al (2019) Trends in intracranial meningioma incidence in the United States, 2004-2015. Cancer Med 8:6458–6467. https://doi.org/10.1002/cam4.2516
Louis DN, Perry A, Wesseling P et al (2021) The 2021 WHO classification of tumors of the Central Nervous System: a summary. Neuro-Oncol 23:1231–1251. https://doi.org/10.1093/neuonc/noab106
Rogers L, Barani I, Chamberlain M et al (2015) Meningiomas: knowledge base, treatment outcomes, and uncertainties. A RANO review. J Neurosurg 122:4–23. https://doi.org/10.3171/2014.7.JNS131644
Islim AI, Mohan M, Moon RDC et al (2019) Incidental intracranial meningiomas: a systematic review and meta-analysis of prognostic factors and outcomes. J Neurooncol 142:211–221. https://doi.org/10.1007/s11060-019-03104-3
Backer-Grøndahl T, Moen BH, Torp SH (2012) The histopathological spectrum of human meningiomas. Int J Clin Exp Pathol 5:231–242
Hanft S, Canoll P, Bruce JN (2010) A review of malignant meningiomas: diagnosis, characteristics, and treatment. J Neurooncol 99:433–443. https://doi.org/10.1007/s11060-010-0348-9
Zaher A, Abdelbari Mattar M, Zayed DH et al (2013) Atypical meningioma: a study of prognostic factors. World Neurosurg 80:549–553. https://doi.org/10.1016/j.wneu.2013.07.001
Loewenstern J, Shuman W, Rutland JW et al (2019) Preoperative and histological predictors of recurrence and survival in atypical Meningioma after initial gross total resection. World Neurosurg 128:e148–e156. https://doi.org/10.1016/j.wneu.2019.04.069
Chohan MO, Ryan CT, Singh R et al (2018) Predictors of treatment response and survival outcomes in Meningioma recurrence with atypical or anaplastic histology. Neurosurgery 82:824–832. https://doi.org/10.1093/neuros/nyx312
Moreau JT, Hankinson TC, Baillet S, Dudley RWR (2020) Individual-patient prediction of meningioma malignancy and survival using the Surveillance, Epidemiology, and end results database. Npj Digit Med 3:12. https://doi.org/10.1038/s41746-020-0219-5
Morin O, Chen WC, Nassiri F et al (2019) Integrated models incorporating radiologic and radiomic features predict meningioma grade, local failure, and overall survival. Neuro-Oncol Adv 1:vdz011. https://doi.org/10.1093/noajnl/vdz011
Kourou K, Exarchos TP, Exarchos KP et al (2015) Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 13:8–17. https://doi.org/10.1016/j.csbj.2014.11.005
Cruz JA, Wishart DS (2007) Applications of machine learning in cancer prediction and prognosis. Cancer Inf 2:59–77
Yousefi S, Amrollahi F, Amgad M et al (2017) Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Sci Rep 7:11707. https://doi.org/10.1038/s41598-017-11817-6
Karabacak M, Margetis K (2023) A machine learning-based Online Prediction Tool for Predicting Short-Term postoperative outcomes following spinal tumor resections. Cancers 15:812. https://doi.org/10.3390/cancers15030812
Karabacak M, Ozkara BB, Ozturk A et al (2022) Radiomics-based machine learning models for prediction of medulloblastoma subgroups: a systematic review and meta-analysis of the diagnostic test performance. Acta Radiol Stockh Swed 1987:2841851221143496. https://doi.org/10.1177/02841851221143496
Bilimoria KY, Stewart AK, Winchester DP, Ko CY (2008) The National Cancer Data Base: a powerful Initiative to improve Cancer Care in the United States. Ann Surg Oncol 15:683–690. https://doi.org/10.1245/s10434-007-9747-3
Collins GS, Reitsma JB, Altman DG, Moons K (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med 13:1. https://doi.org/10.1186/s12916-014-0241-z
Luo W, Phung D, Tran T et al (2016) Guidelines for developing and reporting machine learning predictive models in Biomedical Research: a multidisciplinary view. J Med Internet Res 18:e323. https://doi.org/10.2196/jmir.5870
Akiba T, Sano S, Yanase T et al (2019) Optuna: A Next-generation Hyperparameter Optimization Framework
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. https://doi.org/10.1613/jair.953
Youden WJ (1950) Index for rating diagnostic tests. Cancer 3:32–35. https://doi.org/10.1002/1097-0142(1950)3:1%3C32::aid-cncr2820030106%3E3.0.co;2-3
Fluss R, Faraggi D, Reiser B (2005) Estimation of the Youden Index and its Associated Cutoff Point. Biom J 47:458–472. https://doi.org/10.1002/bimj.200410135
‘Evaluating diagnostic tests and prediction models’ of the STRATOS initiative, On behalf of Topic Group, Van Calster B, McLernon DJ et al (2019) Calibration: the Achilles heel of predictive analytics. BMC Med 17:230. https://doi.org/10.1186/s12916-019-1466-7
Niculescu-Mizil A, Caruana R (2005) Predicting good probabilities with supervised learning. In: Proceedings of the 22nd international conference on Machine learning - ICML ’05. ACM Press, Bonn, Germany, pp 625–632
Bradley AP (1997) The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit 30:1145–1159. https://doi.org/10.1016/S0031-3203(96)00142-2
Lundberg SM, Lee S-I (2017) A Unified Approach to interpreting model predictions. In: Guyon I, Luxburg UV, Bengio S et al (eds) Advances in neural information Processing Systems. Curran Associates, Inc
Goldstein A, Kapelner A, Bleich J, Pitkin E (2015) Peeking inside the Black Box: visualizing statistical learning with plots of individual conditional expectation. J Comput Graph Stat 24:44–65. https://doi.org/10.1080/10618600.2014.907095
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143:29–36. https://doi.org/10.1148/radiology.143.1.7063747
Barthélemy E, Loewenstern J, Konuthula N et al (2018) Primary management of atypical meningioma: treatment patterns and survival outcomes by patient age. J Cancer Res Clin Oncol 144:969–978. https://doi.org/10.1007/s00432-018-2618-4
Champeaux C, Jecko V, Houston D et al (2019) Malignant meningioma: an International Multicentre Retrospective Study. Neurosurgery 85:E461–E469. https://doi.org/10.1093/neuros/nyy610
Aizer AA, Bi WL, Kandola MS et al (2015) Extent of resection and overall survival for patients with atypical and malignant meningioma: extent of resection and recurrence in Meningioma. Cancer 121:4376–4381. https://doi.org/10.1002/cncr.29639
Anakwenze CP, McGovern S, Taku N et al (2020) Association between Facility volume and overall survival for patients with Grade II Meningioma after Gross Total Resection. World Neurosurg 141:e133–e144. https://doi.org/10.1016/j.wneu.2020.05.030
Yang AI, Mensah-Brown KG, Rinehart C et al (2020) Inequalities in Meningioma Survival: results from the National Cancer Database. Cureus. https://doi.org/10.7759/cureus.7304
Acknowledgements
None.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
Conceptualization, MK, PJ, AC, RS, and KM; Methodology, MK, and KM; Software, MK; Formal Analysis, MK; Data Curation, MK; Writing – Original Draft Preparation, MK, and PJ; Writing – Review & Editing, AC, RS and KM; Visualization, MK; Supervision, RS, and KM; Project Administration, MK, and KM.
Corresponding author
Ethics declarations
Disclaimer for the use of web application
The web application should not be used to guide any clinical decisions, including diagnosis and treatment. The authors make no warranties or representations, express or implied, regarding the accuracy, timeliness, relevance, or utility of the information contained in this tool.
Statement for NCDB use
The Commission on Cancer (CoC) of the American College of Surgeons and the American Cancer Society is the source of the data used herein; none of these institutions have verified and are not responsible for the statistical validity of the data analysis or the conclusions derived by the authors.
Institutional review board statement
This study was deemed exempt from approval by the Icahn School of Medicine at Mount Sinai institutional review board because it involved analysis of deidentified patient data.
Source code
The source code for preprocessing and analyzing the data is available on GitHub (https://github.com/mertkarabacak/NCDB-Meningioma).
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Karabacak, M., Jagtiani, P., Carrasquilla, A. et al. Advancing personalized prognosis in atypical and anaplastic meningiomas through interpretable machine learning models. J Neurooncol 164, 671–681 (2023). https://doi.org/10.1007/s11060-023-04463-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-023-04463-8