Abstract
Purpose
Glioblastoma (GBM) is the most common and malignant primary brain tumor in adults with a median overall survival of only 14.6 months despite aggressive treatment. While immunotherapy has been successful in other cancers, its benefit has been proven elusive in GBM, mainly due to a markedly immunosuppressive tumor microenvironment. SARS-CoV-2 has been associated with the development of a pronounced central nervous system (CNS) inflammatory response when infecting different cells including astrocytes, endothelial cells, and microglia. While SARS-CoV2 entry factors have been described in different tissues, their presence and implication on GBM aggressiveness or microenvironment has not been studied on appropriate preclinical models.
Methods
We evaluated the presence of crucial SARS-CoV-2 entry factors: ACE2, TMPRSS2, and NRP1 in matched surgically-derived GBM tissue, cells lines, and organoids; as well as in human brain derived specimens using immunohistochemistry, confocal pixel line intensity quantification, and transcriptome analysis.
Results
We show that patient derived-GBM tissue and cell cultures express SARS-CoV2 entry factors, being NRP1 the most crucial facilitator of SARS-CoV-2 infection in GBM. Moreover, we demonstrate that, receptor expression remains present in our GBM organoids, making them an adequate model to study the effect of this virus in GBM for the potential development of viral therapies in the future.
Conclusion
Our findings suggest that the SARS-CoV-2 virus entry factors are expressed in primary tissues and organoid models and could be potentially utilized to study the susceptibility of GBM to this virus to target or modulate the tumor microenviroment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is responsible for the Coronavirus disease 2019 (COVID-19) [1]. As the mechanisms of viral damage are beginning to be understood, reports have highlighted how SARS-CoV-2 can interact with the host’s intracellular machinery upon cell infection and produce a significant inflammatory response in parenchymal and immune cells [2,3,4,5].
Previous research suggests that SARS-CoV-2 can be detected in the brain and might preferentially infect specific cell populations of the central nervous system (CNS); however, results have been inconsistent and even discordant [6,7,8,9,10,11,12,13]. A proposed mechanism for infection is viral entry into cells with high expression of viral receptors or entry facilitators: angiotensin-converting enzyme-2 (ACE2), transmembrane serine protease-2 (TMPRSS2), and neuropilin-1 (NRP1) among others [14,15,16,17,18]. ACE2 alone can render cells susceptible to SARS-CoV2 infection, but this is usually expressed at low protein levels in respiratory and olfactory epithelial cells [19]. For this reason, co-factors such as TMPRSS2 and NRP1 have been deemed important for SARS-CoV-2 infection, although the list of factors is growing and not only entry factors are required but also genes that are part of the life cycle and replication machinery of the virus.
Glioblastoma (GBM) is the most common malignant brain tumor with a dismal prognosis and no effective treatment despite multiple research efforts [20]. Studies suggest that GBM is susceptible to neurotropic (e.g. Zika, herpes) and oncolytic virus infection [21, 22]. These viruses are being studied in the preclinical setting as therapies against GBM and as tools to understand GBM biology and the tumor microenvironment (TME). There are only scattered reports on the susceptibility of GBM to infection with the novel SARS-CoV-2 virus, limited to an individual case report [21] and in vitro 2D commercial cultures models [22, 23]. In this work, we obtained surgically derived-human tissue to evaluate the expression of SARS-CoV-2 entry factors and to generate a human-derived organoid model that harbors the 3D tumor microenvironment. Our model allows further exploration of the three-dimensional interactions between the SARS-CoV-2 intrinsic machinery and GBM cells. We show that patient-derived glioblastoma tumors present entry factors for SARS-CoV-2 in a patient-specific manner, and that our organoid model recapitulates SARS-CoV-2 receptor expression, making it an important model to study SARS-CoV-2 in GBM.
Methods
Isolation and culture of glioblastoma stem cells
Human GBM tissues were obtained at the Mayo Clinic in Florida (IRB:16-008485). GBM cells were harvested and isolated as previously described [24,25,26,27]. All cell lines were handled in accordance with the biosafety practices and ethical guidelines of UCONN Health and Mayo Clinic that regulate the use of human cells for research.
Glioblastoma and iPSC derived-cerebral organoid culture
For GBM organoid (GBMO) formation, we utilized the methods established in our lab [28, 29]. Briefly, dissociated single cells were cultured with GBM growth media for 4–7 days. GBM neurospheres were transferred to Matrigel droplets (BD Bioscience, San Jose, CA, USA) by pipetting into cold Matrigel on a sheet of Parafilm with 3 mm dimples. These droplets were allowed to gel at 37 °C and were subsequently removed from the Parafilm. After 4 days of stationary growth, tissue droplets were transferred to a spinning bioreactor containing organoid media including 48% Neurobasal, 48% DMEM/F12, 1% B27 supplement, 0.5% N2 supplement, 1% Glutamax, 0.5% MEM-NEAA, insulin, 2-ME and 1% Antibiotic–Antimycotic. Human iPSCs (hiPSC) and iPSC-derived organoids were generated, as previously described [28, 29].
Matched glioblastoma tumor tissue
Matched GBM paraffin block tissues (newly diagnosed, IDH wildtype samples) were obtained from our pathology archives. Samples were obtained from the same patients that the GBM cell lines were derived from. Human brain and lung tissues were obtained from patients deceased from non-neurological conditions.
Immunohistochemistry
5 µm thick sections were obtained, and endogenous peroxidases were deactivated by incubating in peroxidase blocking for 15 min. The slides were then stained with Anti-ACE2 (Abcam, ab15348), Anti-TMPRSS2 (Millipore MABF2158-100UG), and Anti-NRP1 (Proteintech 60067) antibodies. Slides were scanned in the Aperio AT2® and visualized in Aperio ImageScope Software©.
Immunocytochemistry and quantitative confocal microscopy of matched glioblastoma cells
GBM cells were seeded and stained with Anti-ACE2 (Abcam, Cat# ab15348), Anti-TMPRSS2 (Millipore MABF2158-100UG), and Anti-NRP1 (Proteintech 60067–1-Ig) antibodies. To study the distribution of ACE2, TMPRSS2, and NRP-1, we used Quantitative Confocal Microscopy (QCM) with high resolution parameters combined with stereology [30,31,32,33]. Acquisition parameters were established relative to a negative control. We used 2.5D pixel intensity analysis to determine the relative expression of SARS-Cov2 in each neural or GBM stem cells utilizing the Zeiss ZEN Blue Microscope Software.
Quantitative confocal microscopy of glioblastoma organoids
To quantify cells expressing ACE2, NRP1 and TMPRSS2 in each organoid, three pictures per each stained organoid region were taken as 20 × images using confocal laser scanning microscopy (LSM900, ZEISS, Germany). For each organoid, a region designed as “surface” was defined as the organoid tissue contained from the external boundary of each organoid section until 150 um depth. The remaining internal region (> 150 um deeper) was defined as “core”. The number of ACE2, NRP1, or TMPRSS2 positive cells was normalized by number of nuclei (DAPI) per photograph. Comparison among groups was performed using Student's t-test after assessment of data distribution normality.
Gene expression of SARS-CoV-2 entry factors in normal brain and glioblastoma human tissue from publicly available datasets
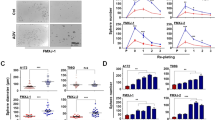
Data regarding transcriptomic expression of ACE2, TMPRSS2, and NRP1 included in Fig. 1A, B were obtained from Zhang Y, et al.[34], and are available at NCBI GEO database: GSE73721. Data in Fig. 1C was obtained from the GTExPortal. Comparison between relative gene expression was performed using one-way ANOVA, Student t-test, or Tukey's multiple comparisons test.
SARS-CoV-2 Entry Facilitators in Normal Huma n Brain. A Mean transcriptomic expression of ACE2, TMPRSS2, and NRP1 after RNA-seq of specific cell cultures isolated from human brains. B Transcriptomic expression of NRP1 according to cell type, comparisons were made using one-way ANOVA and Tukey’s multiple comparisons tests (Astrocytes [GBM periphery] vs. oligodendrocytes (p = 0.0154); Mature astrocytes vs. oligodendrocytes (p = 0.002)) C Heatmap of the transcriptomic expression of ACE2, TMPRSS2, and NRP1 in different regions of normal brain, generated using GTExPortal, transcriptomic expression of NRP1was significantly higher than ACE2 and TMPRSS2 expression (p = 0.0001). D. Immunocytochemistry of SARS-CoV-2 entry factors (ACE2, TMPRSS2 and NRP-1) in patient-derived brain normal tissue. Arrows represent the area that was selected for higher magnification analysis. Astrocytes (white circles), neurons (black circles), oligodendrocytes (blue circles), and ependymal cells (black squares) are identified in all pictures, pictures taken at 20 X magnification
Gliovis®—Data Visualization Tools for Brain Tumor Datasets (http://gliovis.bioinfo.cnio.es/) – was queried to study mRNA expression of SARS-CoV-2 entry factors in GBM and normal brain tissues. Comparisons among different groups (Fig. 2A) were achieved using pairwise comparisons with corrections for multiple testing (p-values with Bonferroni correction).
SARS-CoV-2 Entry Facilitators in Human Glioblastoma. A Transcriptomic expression of ACE2, TMPRSS2 and NRP1 according to RNA-seq data from TCGA in glioblastoma (n = 153) and normal brain (n = 4). Comparisons performed using unpaired t-test after examining for normal distribution. p = ns for ACE2 and TMPRSS2, p = 0.003 for NRP1. B Immunocytochemistry of SARS-CoV-2 entry factors (ACE2, TMPRSS2 and NRP-1) in patient-derived glioblastoma tissue (QNS108, QNS120, and QNS337), pictures were taken at 20 X magnification. C Representative images of a glioblastoma patient-derived cell line (QNS 108) were stained for ACE2 and TMPRSS2, and for ACE2 and NRP-1. All staining was analyzed by Zeiss LSM 800 confocal microscopy. Merged images show co-localization of markers. Scale bars = 10 µm. Graphs at the right show Pixel intensity in QNS108 GBM cells that show distribution of ACE2 (red) and TMPRSS2 (green) and distribution of ACE2 (red) and NRP-1 (green) SARS-COV-2 receptors
Single-cell RNA seq data from human GBM samples was obtained from Müller et al.[35], available at UCSC Cell Browser (https://cells.ucsc.edu/?ds=gbm). Comparisons between percentages of positive populations were performed using Chi-square test. Positive transcriptional expression was defined as a normalized value > 0. Analysis and estimation of tumor infiltration and purity were performed using RNA-seq data of GBM samples available at TCGA as described by Li T, et al.[36, 37], using TIMER2.0 (http://timer.cistrome.org/) and CIBERSORT-ABS and XCELL (for NK cells, CD+4 central memory T cells, and endothelial cells) as methods of analysis. Spearman’s correlation was performed to estimate cell infiltration and gene correlation.
Results
SARS-CoV-2 entry facilitators in healthy human brain
Given the controversy regarding the presence of SARS-CoV-2 entry facilitators in the brain, we explored the expression of ACE2, TMPRSS2, and NRP1 at transcriptional and protein levels. Transcriptional analysis in different types of cells (cortical astrocytes, cortical oligodendrocytes, cortical microglia/macrophages, cortical endothelial cells, cortical neurons, and astrocytes from GBM tumor periphery) from healthy human brains was first carried out. Data was obtained from GEO database: GSE73721 [34]. We found that the transcriptional expression of ACE2 and TMPRSS2 was minimal regardless of the cell type (Fig. 1A). The expression of NRP1 was significantly higher in astrocytes from normal brain and GBM periphery, when compared to oligodendrocytes (p = 0.002 and p = 0.015, respectively) (Fig. 1A, B). A higher transcriptional expression of NRP1 over ACE2 and TMPRSS2 was similarly revealed by analyzing whole tissue of different regions of the brain (p < 0.05) (Fig. 1C).
To confirm these results, we analyzed normal brain tissue from five different patients. Interestingly, despite the low transcriptional expression of ACE2 reported [8], we found a strong expression of ACE2 in most cortical and subcortical astrocytes (white circle). Neurons showed weak positivity in two cases (black circle), and oligodendrocytes (blue circle) were heterogeneously positive in three cases (Fig. 1D). TMPRSS2 expression was positive in the cortex while sparing the white matter, with only weak staining in cortical neurons (black circle) and neuropil but not in other cells of the cortex or white matter. These results correlate with the transcriptomic expression shown in Fig. 1A and C and might reflect the role of other proteins such as NRP1 as more relevant facilitators of SARS-CoV-2 infection in the CNS.NRP1 expression was positive in the cortex. Neurons were positive with nuclear predominance in all samples (black circle). In the white matter, oligodendrocytes were heterogeneously positive to NRP1 in 4 out of 5 patients (blue circle). Astrocytes showed the most robust positivity, with intense staining in the nucleus, cytoplasm, and astrocytic projections (white circle) (Fig. 1D). Interestingly, most NRP1 positive astrocytes were in the cortex or presenting perineural satellitosis. Astrocytic predominance of NRP1 expression is in line with the transcriptomic data presented in Fig. 1B [38]. NRP1 and ACE2 expression was substantial in ependymal cells from all samples (black squares) (Fig. 1D), which is in line with a higher likelihood of SARS-CoV-2 infection in choroid plexus organoids reported by others [12, 13].
SARS-CoV-2 entry facilitators in human glioblastoma
We analyzed the expression of the SARS-CoV-2 entry factors in human GBM. RNA-seq data from 156 GBM tumors and 4 samples of normal brain from the Cancer Genome Atlas Program (TCGA) database were compared (Fig. 2A). Transcriptional expression of ACE2 and TMPRSS2 showed no differences between groups; however, NRP1 expression was significantly higher (p < 0.003) in GBM when compared to normal brain (Fig. 2A).
Furthermore, we used patient-derived GBM tissues and matched cell lines (QNS108, QNS120, QNS140, QNS166, QNS315, and QNS337) and one matched case of tumor recurrence (QNS166-2) (Supplementary Fig. 1). Expression of entry factors had intra- and interpatient heterogeneity, but were positive in all cases (Fig. 2B, C). For tissues, ACE2 was found in the cytoplasm and membrane of GBM tissues and at the dendritic projections of GBM cells. TMPRSS2 presented weak positivity in the cytoplasm and cell membrane and NRP1 expression in GBM tissue was positive in the nucleus, cytoplasm, membrane, and neuropil in all cases (Fig. 2B). Patient-derived cell lines had a positive but heterogeneous signal of all three SARS-CoV-2 entry factors (2C, Supplementary Fig. 3A and B.).
NRP1 is expressed in a cell-type-specific manner in GBM and correlates with an immunosuppressive tumor microenvironment
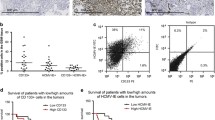
NRP1 is a well-known prognostic factor of survival in GBM and has been described as a critical mediator of SARS-CoV-2 infection [11, 14, 39]. The substantial expression of NRP1 in human GBM tissue samples suggests that NRP1 might play a crucial role as a facilitator of SARS-CoV-2 infection in GBM (Fig. 2). To thoroughly profile the expression of NRP1 in human GBM, we analyzed data from single-cell RNA-seq from eleven GBM patients available at the UCSC Cell Browser [35]. Figure 3A shows the different cell populations identified in human GBM samples (left panel) and their corresponding NRP1 transcriptomic expression (right panel). We found that in GBM cells, NRP1 expression was associated with a specific cell phenotype. Higher expression of NRP1 was identified in cells with mature intermediate proliferating cell (IPC)/Newborn neuron (33.3%) phenotype, radial glia (22.46%), glycolytic progenitor cells (7.84%), and in immature astrocytes (4.85%). Lower NRP1 expression was seen in oligodendrocyte precursor cells (OPC) (1.59%), dividing OPC (0.87%), and neurons (2.76%) (Fig. 3B). Analysis of the tumor microenvironment showed NRP1 mRNA expression was more common in endothelial cells (53.57%), tumor associated macrophages (TAM) (49.1%), pericytes (40%), and microglia (34.61%). B lymphocytes and dividing B lymphocytes were found to express lower percentages of NRP1 positive cells, 0.23% and 0.51%, respectively (Fig. 3C). The elevated transcriptomic expression of NRP1 in intratumoral microglia and TAM and minimal expression in B lymphocytes are in line with the protein expression profile found in peripheral immune circulating cell populations [40]. These results prompted the evaluation of a potential association between NRP1 and the GBM immune tumor microenvironment (TME), which was performed by querying RNA-seq deconvoluted data from 153 GBM patients registered in the TCGA. We found that NRP1 transcriptomic expression was negatively correlated with tumor purity (r = − 0.2, p = 1.88e−2), infiltration of M1 macrophages (r = − 0.202, p = 1.79e−2), natural killer (NK) cells (r = − 0.346, p = 3.4e−5) and CD+ 4 central memory T cells (r = − 0.28, p = 1.05e−3). A positive correlation was found between NRP1 expression and infiltration of M2 macrophages (r = 0.229, p = 7.23e−3), regulatory T cells (r = 0.459, p = 1.73e−8), and endothelial cells (r = 0.288, p = 6.51e−4) (Fig. 3D). Gene ontology enrichment analysis confirmed that leucocyte migration and chemotaxis are among the top 5 most important biological processes mediated by NRP1 protein (Fig. 3E). Taken all together, these results support the idea that GBM is likely to be susceptible to SARS-CoV-2 infection due to the higher expression of the key entry factor NRP1 in GBM cells and the immune cell status in the TME.
Description of NRP1 expression in Glioblastoma and correlation with the Immunosuppressive Tumor Microenvironment. A Transcriptomic expression of NRP1 after single-cell sequencing, each dot represents an individual cell within the different cell groups. B Transcriptomic expression of NRP1 according to glioblastoma cancer cell phenotype. C Transcriptomic expression of NRP1 in Oligodendrocytes and neurons. D Transcriptomic expression of NRP1 in cells of the tumor microenvironment. E Correlation between NRP1 gene expression and tumor purity and infiltration of immune cell in the GBM TME. F Gene ontology enrichment analysis of biological processes associated with NRP1 in GBM
SARS-CoV-2 entry factor expression in human glioblastoma organoids
After we demonstrated the expression of key entry factors of SARS-CoV-2 in human GBM tissue and matched GBM cells, we studied if these factors were also present in GBM organoids (GBMO) (Fig. 4A). To generate an adequate experimental comparison group, human induced pluripotent stem cell (hiPSC)-derived cerebral organoids were also generated and analyzed. We performed immunohistochemistry in GBMO generated from six different human GBM cell lines (GBMO-108, GBMO-120, GBMO-30, GBMO-965, GBMO-1201, and GBMO-640). We co-stained our GBMO with vimentin and, ACE2, TMPRSS2, or NRP1. We found that these cells retained the positivity for ACE2, TMPRSS2, and NRP1 in all GBMOs.
Glioblastoma organoids as an Experimental Model to Study SARS-CoV-2 Infection. A Diagram for glioblastoma organoids (GBMO) fabrication from glioma stem cell. Different from others, our protocol utilizes a successful growth factor-free culture method (left). Pictures at different time point during organoid culture (right). B Immunocytochemistry of SARS-CoV-2 entry factors (ACE2, TMPRSS2 and NRP-1) in patient-derived glioblastoma organoids (QNS108 and GBM-30). Distance between surface and inferior dotted line is 150 um. C Immunocytochemistry of SARS-CoV-2 entry factors (ACE2, TMPRSS2 and NRP-1) in hiPSC-derived cerebral organoid. Distance between surface and inferior dotted line is 150 um. D Quantification of cells positive to ACE2, NRP1 or TMPRSS2 in each organoid
Our group has previously demonstrated the presence of different regions in GBMOs, which are directly related to tumor cell oxygenation and distance from surface [26]. To analyze if the expression of SARS-CoV-2 markers varies upon the distance from surface, we divided the sectional areas of our organoids in “surface” (0–150 um from surface) and “core” (> 150 um from surface) regions. We found the expression of ACE 2 was homogenous across these two different regions, and it was expressed to a lesser extend in the core region of the iPSC-derived organoid (p = 0.0095) (Fig. 4D, Supplementary Fig. 4). TMPRSS expression presented a similar pattern, except from the striking difference found in GBMO-120 surface and core regions (p = 0.0003) (Fig. 4D, Supplementary Fig. 4). The expression of NRP1 seemed to be more associated with the distance from surface, three out of out six GBMOs showed significant differences in the expression levels of NRP1 (GBMO-965, p = 0.0175; GBMO-1201, p = 0.0395; GBMO-640, p = 0.0166), which was also found in our iPSC-derived organoid (p = 0.0494) (Fig. 4B, Supplementary Fig. 4). In all cases where differences were found in SARS-CoV-2 entry factors expression, the “surface” region contained a higher number of positive cells than the “core” region.
Discussion
In this work, we aimed to study the presence of SARS-CoV-2 entry factors in GBM tissue and cell lines. Previous research evaluating SARS-CoV-2 infection in cerebral organoids has shown heterogeneous changes in different types of CNS cell populations, [6,7,8,9, 11], neural progenitors [8], glial cells [10, 11], and ependymal cells of choroid plexus [12, 13]. Our analysis in healthy human brain tissue supports the heterogeneous rates of SARS-CoV-2 infection as we found different expression levels of SARS-CoV-2 entry factors in the different cell populations, with significant interpatient heterogeneity which might be associated with each individual’s genetic background. Despite this heterogeneity, some commonalities exist among patients. For instance, oligodendrocytes have the lowest entry factor expression, while astrocytes presented a strong expression of ACE2 and NRP1. Interestingly, we noticed that astrocytes near neurons (satellite) were more commonly positive to NRP1 than those more distantly located, a phenomenon known as perineural satellitosis. This phenomenon reflects the interaction between astrocytes and neurons and might be related to the astrocytic role as facilitator of neurotropic CNS viral infections [41]. At the same time, both observations correlate with the experimental results from Wang et al., which showed higher SARS-CoV-2 neuronal infectivity when astrocytes are present in culture [19].
While evaluating the presence of SARS-CoV-2 entry factors in GBM and GBMOs [42,43,44], we found a substantial protein expression of ACE2, despite the minimal transcriptomic expression. This discordance between ACE2 transcriptional and protein profiles has also been described in cerebral organoids [11, 45]. TMPRSS2 also presented minimal transcriptional and protein expression, but it was homogeneously positive across all GBM tumor samples, all GBM cell lines, and GBMOs. Recently, Lam et al. demonstrated that the existence of discordant transcriptional and proteomic profiles is common in GBM and suggested this discordance may represent the product of post-transcriptional regulations [46].
NRP1 was strongly expressed in normal brain and GBM at both transcriptomic and proteomic levels, which leads to the hypothesis that NRP1 might be a crucial potentiator of SARS-CoV-2 infection in normal brain [14] and in GBM. Single-cell RNA seq data showed that NRP1 was most expressed in GBM cells and some TME cells such as microglia, TAM, pericytes, and endothelial cells, which further correlated with tumor infiltration of immune cells associated with immunosuppression. The association we found between NRP1 and infiltration of immunosuppressive immune cells also invite the idea of utilizing SARS-CoV-2 as a potential tool to be used for modulation of GBM tumor microenvironment.
We demonstrate that our GBMO harbor a diverse population of GBM cells with different phenotypes and a specific tumor microenvironment which is characterized by a mixed population of proliferative, apoptotic, hypoxic, and quiescent cells [26]. Importantly, by analyzing the transcriptomic profile of our patient derived GBMOs. In previous work we show that each one of them recapitulates different gene networks proper of GBM tumors and share a common intrinsic immune-like genetic program with enrichment of genes involved in cytokine production, antigen processing and presentation, T-cell receptor inhibition, and interferon regulation [26].
Given that non-immune cells GBM cells and tumor-associated astrocytes have proven to be important in regulating the immune tumor microenvironment [47,48,49], GBM organoid models could be a valuable tool to test future hypotheses. Organoid models were previously used to study the effects of different neurotropic viruses [21, 22]; however, there is a lack of reports on the use of organoids to study the effect of SARS-CoV-2 on GBM.
Conclusion
Our data shows that SARS-CoV-2 entry factors, ACE2, TMPRSS2, and NRP1, are positively expressed in patient-derived GBM tissue and matched primary cell cultures, and organoids. Expression of these factors is patient- and cell-type-dependent, being NRP1 the most consistently expressed throughout all our analyses and samples. Moreover, our patient-derived GBM organoids represent an appealing model to study SARS-CoV-2 interactions with GBM. Finally, identifying the expression of SARS-CoV-2 entry factors in GBM supports the rationale of studying the relationship between these two for potential future uses in the clinical setting.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
University, J.H. Coronavirus Resource Center (2021) https://coronavirus.jhu.edu/. Accessed 28 June 2021
Azkur AK et al (2020) Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 75(7):1564–1581
Conti P et al (2020) Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J Biol Regul Homeost Agents 34(2):327–331
Gordon DE et al (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583(7816):459–468
Yang L et al (2020) A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 27(1):125-136.e7
Tiwari SK et al (2021) Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell-derived lung and cerebral organoids. Stem Cell Rep 16(3):437–445
Ramani A et al (2020) SARS-CoV-2 targets neurons of 3D human brain organoids. Embo J 39(20):e106230
Song E et al (2021) Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med 218(3):e20202135
Yi SA et al (2020) Infection of brain organoids and 2D cortical neurons with SARS-CoV-2 pseudovirus. Viruses 12(9):1004
McMahon CL et al (2021) SARS-CoV-2 targets glial cells in human cortical organoids. Stem Cell Rep 16(5):1156–1164
Wang C et al (2021) ApoE-isoform-dependent SARS-CoV-2 neurotropism and cellular response. Cell Stem Cell 28(2):331-342.e5
Jacob F et al (2020) Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell 27(6):937-950.e9
Pellegrini L et al (2020) SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell 27(6):951-961.e5
Cantuti-Castelvetri L et al (2020) Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370(6518):856
Varma P et al (2021) Novel targets of SARS-CoV-2 spike protein in human fetal brain development suggest early pregnancy vulnerability. Front Neurosci. https://doi.org/10.3389/fnins.2020.614680
Samuel RM et al (2020) Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell 27(6):876-889.e12
Hoffmann M et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271-280.e8
Shang J et al (2020) Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA 117(21):11727–11734
Hikmet F et al (2020) The protein expression profile of ACE2 in human tissues. Mol Syst Biol 16(7):e9610
Cantrell JN et al (2019) Progress toward long-term survivors of glioblastoma. Mayo Clin Proc 94(7):1278–1286
Lei J et al (2021) Evidence for residual SARS-CoV-2 in glioblastoma tissue of a convalescent patient. NeuroReport 32(9):771–775
Chu H et al (2020) Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microb 1(1):e14–e23
Bielarz V et al (2021) Susceptibility of neuroblastoma and glioblastoma cell lines to SARS-CoV-2 infection. Brain Res 1758:147344
Guerrero-Cázares H, Chaichana KL, Quiñones-Hinojosa A (2009) Neurosphere culture and human organotypic model to evaluate brain tumor stem cells. Methods Mol Biol 568:73–83
Lara-Velazquez M et al (2021) Alpha 1-antichymotrypsin contributes to stem cell characteristics and enhances tumorigenicity of glioblastoma. Neuro Oncol 23(4):599–610
Watanabe F et al (2021) Patient-derived organoids recapitulate intrinsic immune landscapes and progenitor populations of glioblastoma. biorxiv. https://doi.org/10.1101/2021.10.06.463228
Garcia CA et al (2021) Functional characterization of brain tumor-initiating cells and establishment of gbm preclinical models that incorporate heterogeneity, therapy, and sex differences. Mol Cancer Ther 20(12):2585–2597
Hester ME, Hood AB, (2017) Generation of Cerebral Organoids Derived from Human Pluripotent Stem Cells, in Stem Cell Technologies in Neuroscience, In: Srivastava AK, Snyder EY, and Teng YD (Eds) Springer New York, New York pp. 123–134
Fair SR et al (2020) Electrophysiological maturation of cerebral organoids correlates with dynamic morphological and cellular development. Stem Cell Rep 15(4):855–868
Imitola J et al (2011) Multimodal coherent anti-Stokes Raman scattering microscopy reveals microglia-associated myelin and axonal dysfunction in multiple sclerosis-like lesions in mice. J Biomed Opt 16(2):021109
Rasmussen S et al (2011) Reversible neural stem cell niche dysfunction in a model of multiple sclerosis. Ann Neurol 69(5):878–891
Pluchino S et al (2008) Persistent inflammation alters the function of the endogenous brain stem cell compartment. Brain 131(Pt 10):2564–2578
Wang Y et al (2008) Paradoxical dysregulation of the neural stem cell pathway sonic hedgehog-Gli1 in autoimmune encephalomyelitis and multiple sclerosis. Ann Neurol 64(4):417–427
Zhang Y et al (2016) Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89(1):37–53
Müller S et al (2018) A single-cell atlas of human glioblastoma reveals a single axis of phenotype in tumor-propagating cells. biorxiv 17:377606
Li T et al (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48(W1):W509–W514
Li T et al (2017) TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 77(21):e108–e110
Davies J et al (2020) Neuropilin-1 as a new potential SARS-CoV-2 infection mediator implicated in the neurologic features and central nervous system involvement of COVID-19. Mol Med Rep 22(5):4221–4226
Sun S et al (2017) Neuropilin-1 is a glial cell line-derived neurotrophic factor receptor in glioblastoma. Oncotarget 8(43):74019–74035
Atlas THP (2022) https://www.proteinatlas.org/. Accessed 24 Jan 2022
Potokar M, Jorgačevski J, Zorec R (2019) Astrocytes in Flavivirus Infections. Int J Mol Sci 20(3):691
Zhou Z et al (2021) Multiple strategies to improve the therapeutic efficacy of oncolytic herpes simplex virus in the treatment of glioblastoma. Oncol Lett 22(1):510
Zhu Z et al (2017) Zika virus has oncolytic activity against glioblastoma stem cells. J Exp Med 214(10):2843–2857
Zhu Z et al (2020) Zika virus targets glioblastoma stem cells through a SOX2-Integrin α(v)β(5) Axis. Cell Stem Cell 26(2):187-204.e10
Song E et al (2021) Divergent and self-reactive immune responses in the CNS of COVID-19 patients with neurological symptoms. Cell Rep Med 2(5):100288
Lam KHB et al (2022) Topographic mapping of the glioblastoma proteome reveals a triple-axis model of intra-tumoral heterogeneity. Nat Commun 13(1):116
Henrik Heiland D et al (2019) Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat Commun 10(1):2541
Wang Q et al (2017) Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32(1):42-56.e6
Wu A et al (2010) Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol 12(11):1113–1125
Funding
This work was supported by the following grants: Distinguished Mayo Clinic Investigator Award (AQH), the William J and Charles H Mayo Professor (AQH), the Uihlein Neuro-Oncology Research Fund (AQH), and NIH (R01CA216855 and R33CA240181).
Author information
Authors and Affiliations
Contributions
Conception and design: PSM, FW, HRG, PS, JI, AQH. Financial Support: JI, AQH. Administrative Support: PS, JI, AQH. Provision of Study Material of Patients: MJ, JI, AQH. Collection and/or Assembly of Data: PSM, FW, HRG, SR, DG, PS, JI, AHQ. Data analysis and interpretation: all authors. Manuscript Writing: PSM, FW, HRG. Final Approval of Manuscript: all authors.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Suarez-Meade, P., Watanabe, F., Ruiz-Garcia, H. et al. SARS-CoV2 entry factors are expressed in primary human glioblastoma and recapitulated in cerebral organoid models. J Neurooncol 161, 67–76 (2023). https://doi.org/10.1007/s11060-022-04205-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-022-04205-2