Abstract
Introduction
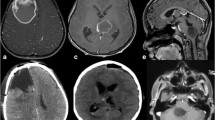
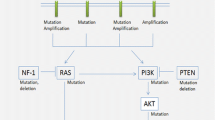
Glioblastoma (GBM) is the most aggressive central nervous system (CNS) tumor with astrocytic differentiation. The growth pattern of GBM mimics that of the precursor cell migration during the fetal development of the brain. Diaphanous homolog (Diaph3) has been established to play a role in both CNS maturation and cancer progression as it is required both for cell migration and division. Furthermore, Diaph3 has been shown to play a role in malignant disease progression through hyperactivation of the EGFR/MEK/ERK in loss of expression and its overexpression correlating to hyperactivity of the mTOR pathway, both of which are with a well-established role in GBM. Herein, we aimed at establishing the diagnostic role of Diaph3 immunohistochemistry expression patterns in GBM and their possible implications for molecular response to different therapies.
Materials and methods
The study utilized a retrospective nonclinical approach. Results of Diaph3 immunohistochemical expression were compared to healthy controls and reactive gliosis and statistically analyzed for correlation with neuroradiological tumor parameters and patient survival.
Results
Healthy controls showed individual weakly positive cells, while reactive gliosis controls showed a strong expression in astrocytic projections. GBM samples showed a heterogeneous positive reaction to Diaph3, mean number of positive cells 62.66%, median 61.5, range 12–96%. Areas of migrating cells showed a strong diffuse cytoplasmic reaction. Cells located in the tumor core and those in areas of submeningeal aggregation had no antibody expression. Statistical analysis revealed no correlation with tumor size or patient survival.
Conclusion
The different expression pattern of Diaph3 in healthy controls, reactive gliosis and GBM shows promise as a clinical differentiating marker. Despite Diaph3 expression not correlating with survival and tumor size in GBM, there is an accumulating body of evidence that Diaph3 correlates with mTOR activity and can thus be used as a predictor for response to rapamycin and taxanes, clinical studies of which have shown promising, if mixed results in GBM.




Similar content being viewed by others
References
Louis DN, Perry A, Wesseling P et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23:1231–1251
Brat DJ, Aldape K, Colman H et al (2020) cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol 139:603–608
Volovetz J, Berezovsky AD, Alban T et al (2020) Identifying conserved molecular targets required for cell migration of glioblastoma cancer stem cells. Cell Death Dis 11:1–12
Tamimi AF, Juweid M (2017) Epidemiology and Outcome of Glioblastoma. In: De Vleeschouwer S (ed) Glioblastoma. Codon Publications, Brisbane, pp 143–153
Hara A, Kanayama T, Noguchi K et al (2019) Treatment strategies based on histological targets against invasive and resistant glioblastoma. J Oncol 2019:1–10
Gillespie S, Monje M (2018) An active role for neurons in glioma progression: making sense of Scherer’s structures. Neuro Oncol 20:1292–1299
Wesseling P, Kros JM, Jeuken JWM (2011) The pathological diagnosis of diffuse gliomas: towards a smart synthesis of microscopic and molecular information in a multidisciplinary context. Diagnostic Histopathol 17:486–494
Habberstad AH, Habberstad AH, Lind-Landström T, Torp SH (2012) The histopathological spectrum of primary human glioblastomas with relations to tumour biology. J Clin Exp Pathol 2:1–7
Zepecki JP, Snyder KM, Moreno MM et al (2018) Regulation of human glioma cell migration, tumor growth, and stemness gene expression using a Lck targeted inhibitor. Oncogene 38:1734–1750
Munthe S, Sørensen MD, Thomassen M et al (2016) Migrating glioma cells express stem cell markers and give rise to new tumors upon xenografting. J Neurooncol 130:53–62
Holland EC (2000) Glioblastoma multiforme: the terminator. Proc Natl Acad Sci USA 97:6242
Wang CH, Rockhill JK, Mrugala M et al (2009) Prognostic significance of growth kinetics in newly diagnosed glioblastomas revealed by combining serial imaging with a novel biomathematical model. Cancer Res 69:9133–9140
Boaro A, Harary M, Chukwueke U, Valdes Quevedo P, Smith TR (2019) The neurocognitive evaluation in the butterfly glioma patient. A systematic review. Interdiscip Neurosurg 18:100512
Tunthanathip T, Ratanalert S, Sae-Heng S, Oearsakul T (2017) Butterfly tumor of the corpus callosum: Clinical characteristics, diagnosis, and survival analysis. J Neurosci Rural Pract 8:S57
Cacho-Díaz B, García-Botello DR, Wegman-Ostrosky T, Reyes-Soto G, Ortiz-Sánchez E, Herrera-Montalvo LA (2020) Tumor microenvironment differences between primary tumor and brain metastases. J Transl Med 18:1–12
Patnayak R, Jena A, Vijaylaxmi B et al (2013) Metastasis in central nervous system: Clinicopathological study with review of literature in a tertiary care center in South India. South Asian J Cancer 2:245–249
Damiani D, Goffinet AM, Alberts A, Tissir F (2016) Lack of Diaph3 relaxes the spindle checkpoint causing the loss of neural progenitors. Nat Commun 7:1–12
DIAPH3 Gene - GeneCards | DIAP3 Protein | DIAP3 Antibody (2022) Available from: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DIAPH3. Accessed Jan 15 2022.
Kawabata Galbraith K, Kengaku M (2019) Multiple roles of the actin and microtubule-regulating formins in the developing brain. Neurosci Res 138:59–69
Dong L, Li Z, Xue L et al (2018) DIAPH3 promoted the growth, migration and metastasis of hepatocellular carcinoma cells by activating beta-catenin/TCF signaling. Mol Cell Biochem 438:183–190
Hager MH, Morley S, Bielenberg DR et al (2012) DIAPH3 governs the cellular transition to the amoeboid tumour phenotype. EMBO Mol Med 4:743–760
Morley S, You S, Pollan S et al (2015) Regulation of microtubule dynamics by DIAPH3 influences amoeboid tumor cell mechanics and sensitivity to taxanes. Sci Reports 5:1–16
Pettee KM, Becker KN, Alberts AS, Reinard KA, Schroeder JL, Eisenmann KM (2019) Targeting the mDia formin-assembled cytoskeleton is an effective anti-invasion strategy in adult high-grade glioma patient-derived neurospheres. Cancers 11:392
Arden JD, Lavik KI, Rubinic KA et al (2015) Small-molecule agonists of mammalian diaphanous–related (mDia) formins reveal an effective glioblastoma anti-invasion strategy. Mol Biol Cell 26:3704–3718
Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei M (2008) Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol Biol Evol 25:2717–2733
Evangelista M, Zigmond S, Boone C (2003) Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci 116:2603–2611
Schönichen A, Geyer M (2010) Fifteen formins for an actin filament: A molecular view on the regulation of human formins. Biochim Biophys Acta - Mol Cell Res 1803:152–163
Labat-de-hoz L, Alonso MA (2021) Formins in human disease. Cells 10:2554
Palander O, Lam A, Collins RF, Moraes TJ, Trimble WS (2021) Nonredundant roles of DIAPHs in primary ciliogenesis. J Biol Chem 296:100680
Lau EOC, Damiani D, Chehade G et al (2021) DIAPH3 deficiency links microtubules to mitotic errors, defective neurogenesis, and brain dysfunction. Elife 10:e61974
Vorstman JAS, Van Daalen E, Jalali GR et al (2011) A double hit implicates DIAPH3 as an autism risk gene. Mol Psychiatry 16:442–451
Shun WJ, Jiang J, Jun CB, Wang K, Ling TY, Xhua L (2021) Plasticity of cancer cell invasion: Patterns and mechanisms. Transl Oncol 14:100899
Wan L, Zhu J, Wu Q (2021) Knockdown of DIAPH3 inhibits the proliferation of cervical cancer cells through inactivating mTOR signaling pathway. J Oncol 2021:1–16
Read RD, Cavenee WK, Furnari FB, Thomas JB (2009) A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet 5:e1000374
Gulati N, Karsy M, Albert L, Murali R, Jhanwar-Uniyal M (2009) Involvement of mTORC1 and mTORC2 in regulation of glioblastoma multiforme growth and motility. Int J Oncol 35:731–740
Liberti MV, Locasale JW (2016) The warburg effect: how does it benefit cancer cells? Trends Biochem Sci 41:211–218
Mecca C, Giambanco I, Donato R, Arcuri C (2018) Targeting mTOR in glioblastoma: rationale and preclinical/clinical evidence. Dis Markers 2018:9230479
Tanaka K, Babic I, Nathanson D et al (2011) Oncogenic EGFR signaling activates an mTORC2-NF-κB pathway that promotes chemotherapy resistance. Cancer Discov 1:524–538
Chandrika G, Natesh K, Ranade D, Chugh A, Shastry P (2016) Suppression of the invasive potential of Glioblastoma cells by mTOR inhibitors involves modulation of NFκB and PKC-α signaling. Sci Reports 6:1–14
Heimberger AB, Wang E, McGary EC et al (2005) Mechanisms of action of rapamycin in gliomas. Neuro Oncol 7:1–11
Zimmerman MA, Wilkison S, Qi Q, Chen G, Li PA (2020) Mitochondrial dysfunction contributes to rapamycininduced apoptosis of human glioblastoma cells - a synergistic effect with temozolomide. Int J Med Sci 17:2831–2843
Arcella A, Biagioni F, Antonietta Oliva M et al (2013) Rapamycin inhibits the growth of glioblastoma. Brain Res 1495:37–51
Cloughesy TF, Yoshimoto K, Nghiemphu P et al (2008) Antitumor activity of rapamycin in a phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med 5:e8
Li M, Zhou Y, Chen C et al (2019) Efficacy and safety of mTOR inhibitors (rapamycin and its analogues) for tuberous sclerosis complex: a meta-analysis. Orphanet J Rare Dis 14:1–9
Funding
The study was funded by the Medical University – Varna Scientific fund, grant number 19010 and the National scientific fund—young researchers, grant number 2990/07.06.2021.
Author information
Authors and Affiliations
Contributions
GSS conceptualized the study, analyzed preliminary findings, performed the statistical analysis and wrote the initial draft with figure design; EL collected patient data, revised findings and statistical results; RG analyzed preliminary findings and assisted with statistical analysis and figure design; DD and LP assisted in patient selection and data analysis; AK and BDI assisted in statistical analysis and finding interpretation; PG revised the findings, manuscript and figures as well as approved the final version of the manuscript; all althours have read and approve the final version of the manuscript
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Ethical approval
All procedures included in the study adhered to the ethical standards of the Declaration of Helsinki 1964 and its seventh revision from 2013. The study was approved by the Committee on Ethics for Scientific Research, Medical University—Varna "Prof. Dr. Paraskev Stoyanov," —Protocol no. 93/21.05.2020.
Conflict of interest
The authors declare no conflicting interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Stoyanov, G.S., Lyutfi, E., Georgieva, R. et al. Diaph3 underlines tumor cell heterogeneity in glioblastoma with implications for treatment modalities resistance. J Neurooncol 157, 523–531 (2022). https://doi.org/10.1007/s11060-022-03996-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-022-03996-8