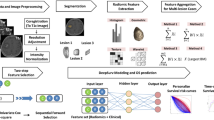
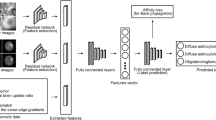
Abstract
Purpose
Non-small cell lung cancer (NSCLC) tends to metastasize to the brain. Between 10 and 60% of NSCLCs harbor an activating mutation in the epidermal growth-factor receptor (EGFR), which may be targeted with selective EGFR inhibitors. However, due to a high discordance rate between the molecular profile of the primary tumor and the brain metastases (BMs), identifying an individual patient’s EGFR status of the BMs necessitates tissue diagnosis via an invasive surgical procedure. We employed a deep learning (DL) method with the aim of noninvasive detection of the EGFR mutation status in NSCLC BM.
Methods
We retrospectively collected clinical, radiological, and pathological-molecular data of all the NSCLC patients who had been diagnosed with BMs and underwent resection of their BM during 2009–2019. The study population was then divided into two groups based upon EGFR mutational status. We further employed a DL technique to classify the two groups according to their preoperative magnetic resonance imaging features. Augmentation techniques, transfer learning approach, and post-processing of the predicted results were applied to overcome the relatively small cohort. Finally, we established the accuracy of our model in predicting EGFR mutation status of BM of NSCLC.
Results
Fifty-nine patients were included in the study, 16 patients harbored EGFR mutations. Our model predicted mutational status with mean accuracy of 89.8%, sensitivity of 68.7%, specificity of 97.7%, and a receiver operating characteristic curve value of 0.91 across the 5 validation datasets.
Conclusion
DL-based noninvasive molecular characterization is feasible, has high accuracy and should be further validated in large prospective cohorts.


Similar content being viewed by others
Data availability
All the data in this study is available in Sagol Brain Institute, Tel Aviv Medical Center, Tel-Aviv, Israel.
Code availability
Not applicable.
References
Bray F, Ferlay J, Soerjomataram I et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21492
Molina JR, Yang P, Cassivi SD et al (2008) Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 83:584–594. https://doi.org/10.4065/83.5.584
Schouten LJ, Rutten J, Huveneers HAM, Twijnstra A (2002) Incidence of brain metastases in a cohort of patients with carcinoma of the breast, colon, kidney, and lung and melanoma. Cancer 94:2698–2705. https://doi.org/10.1002/cncr.10541
Chi A, Komaki R (2010) Treatment of brain metastasis from lung cancer. Cancers 2:2100–2137. https://doi.org/10.3390/cancers2042100
Shin D-Y, Na II, Kim CH et al (2014) EGFR mutation and brain metastasis in pulmonary adenocarcinomas. J Thorac Oncol 9:195–199. https://doi.org/10.1097/JTO.0000000000000069
Ali A, Goffin JR, Arnold A, Ellis PM (2013) Survival of patients with non-small-cell lung cancer after a diagnosis of brain metastases. Curr Oncol Tor Ont 20:e300-306. https://doi.org/10.3747/co.20.1481
Oda K, Matsuoka Y, Funahashi A, Kitano H (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 1:E1–E17. https://doi.org/10.1038/msb4100014
Midha A, Dearden S, McCormack R (2015) EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 5:2892–2911
Mok TS, Wu Y-L, Thongprasert S et al (2009) Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361:947–957. https://doi.org/10.1056/NEJMoa0810699
Rosell R, Carcereny E, Gervais R et al (2012) Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 13:239–246. https://doi.org/10.1016/S1470-2045(11)70393-X
Sequist LV, Yang JC-H, Yamamoto N et al (2013) Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 31:3327–3334. https://doi.org/10.1200/JCO.2012.44.2806
Soria J-C, Ohe Y, Vansteenkiste J et al (2018) Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 378:113–125. https://doi.org/10.1056/NEJMoa1713137
Kalikaki A, Koutsopoulos A, Trypaki M et al (2008) Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer 99:923. https://doi.org/10.1038/sj.bjc.6604629
Bozzetti C, Tiseo M, Lagrasta C et al (2008) Comparison between epidermal growth factor receptor (EGFR) gene expression in primary non-small cell lung cancer (NSCLC) and in fine-needle aspirates from distant metastatic sites. J Thorac Oncol 3:18–22. https://doi.org/10.1097/JTO.0b013e31815e8ba2
Brastianos PK, Carter SL, Santagata S et al (2015) Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov 5:1164–1177. https://doi.org/10.1158/2159-8290.CD-15-0369
Berger LA, Riesenberg H, Bokemeyer C, Atanackovic D (2013) CNS metastases in non-small-cell lung cancer: current role of EGFR-TKI therapy and future perspectives. Lung Cancer 80:242–248. https://doi.org/10.1016/j.lungcan.2013.02.004
Shofty B, Artzi M, Shtrozberg S et al (2020) Virtual biopsy using MRI radiomics for prediction of BRAF status in melanoma brain metastasis. Sci Rep 10:6623. https://doi.org/10.1038/s41598-020-63821-y
Quon JL, Bala W, Chen LC et al (2020) Deep learning for pediatric posterior fossa tumor detection and classification: a multi-institutional study. Am J Neuroradiol 41:1718–1725. https://doi.org/10.3174/ajnr.A6704
Lundervold AS, Lundervold A (2019) An overview of deep learning in medical imaging focusing on MRI. Z Für Med Phys 29:102–127. https://doi.org/10.1016/j.zemedi.2018.11.002
Litjens G, Kooi T, Bejnordi BE et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88. https://doi.org/10.1016/j.media.2017.07.005
Grossman R, Haim O, Abramov S et al (2021) Differentiating small-cell lung cancer from non-small-cell lung cancer brain metastases based on MRI using efficientnet and transfer learning approach. Technol Cancer Res Treat 20:15330338211004920. https://doi.org/10.1177/15330338211004919
Ahn SJ, Kwon H, Yang J-J et al (2020) Contrast-enhanced T1-weighted image radiomics of brain metastases may predict EGFR mutation status in primary lung cancer. Sci Rep. https://doi.org/10.1038/s41598-020-65470-7
Park YW, An C, Lee J et al (2021) Diffusion tensor and postcontrast T1-weighted imaging radiomics to differentiate the epidermal growth factor receptor mutation status of brain metastases from non-small cell lung cancer. Neuroradiology 63:343–352. https://doi.org/10.1007/s00234-020-02529-2
Scrivener M, de Jong EEC, van Timmeren JE et al (2016) Radiomics applied to lung cancer: a review. Transl Cancer Res. https://doi.org/10.21037/8536
Li Q, Bai H, Chen Y et al (2017) A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep 7:14331. https://doi.org/10.1038/s41598-017-14753-7
Sun Q, Lin X, Zhao Y et al (2020) Deep learning vs. radiomics for predicting axillary lymph node metastasis of breast cancer using ultrasound images: don’t forget the peritumoral region. Front Oncol. https://doi.org/10.3389/fonc.2020.00053
Ashburner J, Friston KJ (2005) Unified segmentation. Neuroimage 26:839–851. https://doi.org/10.1016/j.neuroimage.2005.02.018
Howard J, Gugger S (2020) Fastai: a layered API for deep learning. Information 11:108. https://doi.org/10.3390/info11020108
Zhang H, Cisse M, Dauphin YN, Lopez-Paz D (2018) mixup: beyond empirical risk minimization. ArXiv171009412 Cs Stat
Liu W, Anguelov D, Erhan D et al (2016) SSD: Single shot multibox detector. In: Leibe B, Matas J, Sebe N, Welling M (eds) Computer vision – ECCV 2016. Springer International Publishing, Cham, pp 21–37
Iglovikov V, Shvets A (2018) TernausNet: U-Net with VGG11 encoder pre-trained on imagenet for image segmentation. ArXiv180105746 Cs
Lau SLH, Chong EKP, Yang X, Wang X (2020) Automated pavement crack segmentation using U-Net-based convolutional neural network. IEEE Access 8:114892–114899. https://doi.org/10.1109/ACCESS.2020.3003638
Tajbakhsh N, Shin JY, Gurudu SR et al (2016) Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 35:1299–1312. https://doi.org/10.1109/TMI.2016.2535302
Raghu M, Zhang C, Kleinberg J, Bengio S (2019) Transfusion: understanding transfer learning for medical imaging. In: Thrun S, Saul LK, Schölkopf B (eds) Advances in neural information processing systems. Curran Associates Inc, Red Hook
Frid-Adar M, Ben-Cohen A, Amer R, Greenspan H (2018) Improving the Segmentation of anatomical structures in chest radiographs using U-Net with an Imagenet pre-trained encoder. In: Third International Workshop, RAMBO 2018, Fourth International Workshop, BIA 2018, and First International Workshop, TIA 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16 and 20, 2018, Proceedings. pp 159–168
Chen B, Zhang R, Gan Y et al (2017) Development and clinical application of radiomics in lung cancer. Radiat Oncol 12:154. https://doi.org/10.1186/s13014-017-0885-x
Oikonomou A, Khalvati F, Tyrrell PN et al (2018) Radiomics analysis at PET/CT contributes to prognosis of recurrence and survival in lung cancer treated with stereotactic body radiotherapy. Sci Rep 8:4003. https://doi.org/10.1038/s41598-018-22357-y
Hosny A, Parmar C, Coroller TP et al (2018) Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med 15:e1002711. https://doi.org/10.1371/journal.pmed.1002711
Trebeschi S, Drago SG, Birkbak NJ et al (2019) Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol Off J Eur Soc Med Oncol 30:998–1004. https://doi.org/10.1093/annonc/mdz108
Ortiz-Ramón R, Larroza A, Ruiz-España S et al (2018) Classifying brain metastases by their primary site of origin using a radiomics approach based on texture analysis: a feasibility study. Eur Radiol. https://doi.org/10.1007/s00330-018-5463-6
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
OH, BS, ZR, MA and RG contributed to the study conception and design. Material preparation and data collection were performed by SA, OH and CF, analysis was performed by MA and NA. The first draft of the manuscript was written by OH and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
Tel-Aviv Medical Center, and Fondazio ne IRCCS Istituto Neurologico C. Besta, (IRB approval numbers 0200-10, and 81/2021, respectively).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Haim, O., Abramov, S., Shofty, B. et al. Predicting EGFR mutation status by a deep learning approach in patients with non-small cell lung cancer brain metastases. J Neurooncol 157, 63–69 (2022). https://doi.org/10.1007/s11060-022-03946-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-022-03946-4