Abstract
Introduction
Medulloblastoma (MB) is an embryonal tumour that originates from genetic deregulation of cerebellar developmental pathways and is classified into 4 molecular subgroups: SHH, WNT, group 3, and group 4. Hydroxymethylation levels progressively increases during cerebellum development suggesting a possibility of deregulation in MB pathogenesis. The aim of this study was to investigate global hydroxymethylation levels and changes in TET and IDH gene expression in MB samples compared to control cerebellum samples.
Methods
The methods utilized were qRT-PCR for gene expression, dot-blot and immunohistochemistry for global hydroxymethylation levels and sequencing for the investigation of IDH mutations.
Results
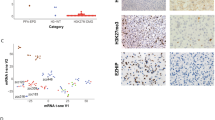
Our results show that global hydroxymethylation level was decreased in MB, and low 5hmC level was associated with the presence of metastasis. TET1 expression levels were decreased in the WNT subgroup, while TET3 expression levels were decreased in the SHH subgroup. Reduced TET3 expression levels were associated with the presence of events such as relapse and death. Higher expression of IDH1 was observed in MB group 3 samples, whereas no mutations were detected in exon 4 of IDH1 and IDH2.
Conclusion
These findings suggest that reduction of global hydroxymethylation levels, an epigenetic event, may be important for MB development and/or maintenance, representing a possible target in this tumour and indicating a possible interaction of TET and IDH genes with the developmental pathways specifically activated in the MB subgroups. These genes could be specific targets and markers for each subgroup.



Similar content being viewed by others
References
Chan AW, Tarbell NJ, Black PM et al (2000) Adult medulloblastoma: prognostic factors and patterns of relapse. Neurosurgery 47:623–631
Crawford JR, Macdonald TJ, Packer RJ (2007) Medulloblastoma in childhood: new biological advances. Lancet Neurol 6:1073–1085
Louis DN, Ohgaki H, Wiestler OD et al (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114:97–109
Packer RJ, Cogen P, Vezina G et al (1999) Medulloblastoma: clinical and biologic aspects. Neuro Oncol 1:232–250
Smoll NR, Drummond KJ (2012) The incidence of medulloblastomas and primitive tumours in adults and children. J Clin Neurosci 19:1541–1544
Mabbot DJ, Penkman L, Witol A et al (2008) Core neurocognitive functions in children treated for posterior fossa tumors. Neuropsychology 22:159–168
Hatten M, Roussel MF (2011) Development and cancer of the cerebellum. Cell Press 34:134–142
Northcott PA, Jones D, Kool M et al (2012) Medulloblastomics: the end of the beginning. Nat Rev 12:818–834
Taylor MD (2012) Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123:465–447
Kongkham PN, Northcott PA, Croul SE et al (2010) The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene 29:3017–3024
Pócza T, Krenács T, Turányi E et al (2016) High expression of DNA methyltransferase in primary human medulloblastoma. Folia Neuropathol 54(2):105–113
Tahiliani M, Koh KP, Shen Y et al (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935
Willians K, Christensen J, Helin K (2012) DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep 1:28–35
Haffner MC, Chaux A, Meeker AK et al (2011) Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2:627–637
Kudo Y, Tateishi K, Yamamoto K et al (2012) Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci 103(4):670–676
Lian CG, Xu Y, Ceol C et al (2012) Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 6:1135–1146
Liu C, Liu L, Chen X et al (2013) Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS ONE 8(5):e62828
Yang H, Liu Y, Bai F et al (2013) Tumor development is associated with decreased of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 32:663–669
Frycz BA, Murawa D, Borejsza-Wysocki M et al (2014) Decreased expression of ten-eleven translocation 1 protein is associated with some clinicopathological features in gastric cancer. Biomed Pharmacother 68:209–212
Dong Z-R, Zhang C, Cai J-B et al (2014) Role of 5-hydroxymethylcytosine level in diagnosis and prognosis prediction of intrahepatic cholangiocarcinoma. Tumor Biol 4:2763–2771
Feng J, Wang Q, Guanguei L et al (2015) TET1 mediated different transcriptional regulation in prostate cancer. Inter J Clin Exp Med 8:203–211
Murata A, Baba Y, Ishimoto T et al (2015) Tet family proteins and 5-hydroxymethylcytosine esophageal squamous cell carcinoma. Oncotarget 6:23372–23382
Chen Z, Shi X, Guo L et al (2017) Decreased 5-hydroxymethylcytosine levels correlate with cancer progression and poor survival: a systematic review and meta-analysis. Oncotarget 8(1):1944–1952
Xu Y, Yang H, Liu Y et al (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarato-dependente dioxygenases. Cancer Cell 19:17–30
Figueroa ME, Abdel-Wahab O, Lu C et al (2010) Leukemia IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 6:553–567. https://doi.org/10.1016/j.ccr.2010.11.015
Yan H, Parsons W, Jin G et al (2010) IDH1 and IDH2 in gliomas. NIH Public Access 8:765–773
Wang T, Pan Q, Lin L et al (2012) Genome-wide DNA hydroxymethylation changes are associated with neurodevelopment genes in the developing human cerebellum. Hum Mol Genet 21:5500–5510
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108
Pearson K (1896) Mathematical contributions to the theory of evolution. III. Regression, heredity, and panmixia. Philos Trans R Soc Lond Ser A 187:253–318
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300
Patnaik MM, Hanson C, Hodnefield JM et al (2012) Differential prognostic effect of IDH1 versus IDH2 mutations in myelodysplastic syndromes: a Mayo Clinic study of 277 patients. Leukemia 26(1):101–105
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A (2012) Geneious Basic: anintegrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Jin SJ, Jiang Y, Qiu R (2011) 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 71(24):7360–7365
Orr B, Haffner MC, Nelson WG et al (2012) Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS ONE 7:e41036
Kraus TFJ, Globisch D, Wagner M et al (2012) Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int J Cancer 7:1577–1590
Tsai K-W, Li G-C, Chen C-H et al (2015) Reduction of global 5-hydroxymethycytosine is a poor factor in breast cancer patients, especially for an ER/PR-negative subtype. Breast Cancer Res Treat 153:219–234
Song C-X, Yin S, Ma L et al (2017) 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res 27(10):1231–1242
Li W, Zhang X, Lu X et al (2017) 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res 27(10):1243–1257
Kafer GR, Li X, Horii T et al (2016). 5-Hydroxymethylcytosine marks sites of DNA damage and promotes genome stability. Cell Rep 14:1–10
Tomkova M, McClellan M, Kriaucionis S et al (2016). 5-hydroxymethylcytosine marks regions with reduced mutation frequency in human DNA. eLife. https://doi.org/10.7554/eLife.17082
Zhou Z, Zhang H-S, Liu Y et al (2017). Loss of TET1 facilitates DLD1 colon cancer cell migration via H3K27me3-mediated down-regulation of E-cadherin. J Cell Physiol. https://doi.org/10.1002/jcp.26012
Neri F, Dettori D, Incarnato D et al (2014) TET1 is a tumour suppressor that inhibits colon cancer growth by repressing inhibitors of the WNT pathway. Oncogene 32:4168–4176. https://doi.org/10.1038/onc.2014.356
Kim R, Sheaffer KL, Choi I et al (2016) Epigenetic regulation of intestinal stem cells by Tet1-mediated DNA hydroxymethylation. Genes Dev 30(21):2433–2442
Duan H, Yan Z, Chen W et al (2017) TET1 inhibits EMT of ovarian cancer cells through activating Wnt/β-catenin signaling inhibitors DKK1 and SFRP2. Gynecol Oncol 147(2):408–417
Schwalbe EC, Lindsey JC, Nakjang S et al (2017) Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol 18(7):958–971
Cui Q, Yang S, Ye P et al (2016). Downregulation of TLX induces TET3 expression and inhibits glioblastoma stem cell self-renewal and tumorigenesis. Nat Commun 7:10637
Perera A, Eisen D, Wagner M et al (2015). TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep 11:283–294
Uribes-Lewis S, Stark R, Carrol T et al (2015) 5-hydroxymethylcytosine marks promoters in colon that resist DNA hypermethylation in cancer. Genome Biol 16:69–84
Balss J, Meyer J, Mueller W et al (2008) Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116:597–602. https://doi.org/10.1007/s00401-008-0455-2
Calvert AE, Chalastanis A, Wu Y et al (2017) Cancer-associated IDH1 promotes growth and resistance to targeted therapies in the absence of mutation. Cell Rep 19(9):1858–1873
Du J, Martin SM, Levine M et al (2013) Mechanisms of ascorbate-induced cytotoxicity in pancreatic cancer. Clin Cancer Res 16(2):509–520
Chen J, Guo L, Zhang L et al (2013) Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet 45(12):1504–1509
Dickson KM, Gustafson CB, Young JI et al (2013) Ascorbate-induced generation of 5-hydroxymethylcytosine is unaffected by varying levels of iron and 2-oxoglutarate. Biochem Biophys Res Commun 439(4):522–527
Minor EA, Court BL, Young JI et al (2013) Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem 288(19):13669–13674
Yin R, Mao S-Q, Zhao B et al (2013) Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc 135(28):10396–10403
Blaschke K, Ebata KT, Karimi MM et al (2013) Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500(7461):222–226
Cieslak J, Cullen JJ et al (2015) Treatment of pancreatic cancer with pharmacological ascorbate. Curr Pharm Biotechnol 16(9):759–770
Gustafson CB, Yang C, Dickson KM et al (2015) Epigenetic reprogramming of melanoma cells by vitamin C treatment. Clin Epigenetics 7(1):51
Funding
This work was funded by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Process Number 2013/15125-3 and 2014/20341-0). Fundação de Apoio ao Ensino, Pesquisa e Assistência (FAEPA) do Hospital das Clínicas da Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This research was submitted to and approved by the HC/FMRP-USP Research Ethics Committee (CAAE n° 37206114.1.0000.5440).
Informed consent
All samples were obtained after receiving informed consent from all participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11060_2018_2845_MOESM1_ESM.tif
Kaplan-Meier curves for event-free survival according to 5hmC levels in MB. A. Using dot-blot (n=11); B. Using IHC (n=30) (TIF 11158 KB)
11060_2018_2845_MOESM2_ESM.tif
Kaplan-Meier curves for event-free survival according to A. TET1 gene expression; B. TET2 gene expression; C. TET3 gene expression; D. IDH1 gene expression; E. IDH2 gene expression in MB samples (TIF 24757 KB)
11060_2018_2845_MOESM3_ESM.tif
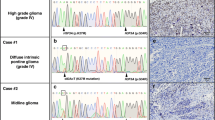
Representative sequencing electropherograms of Isocitrate Dehydrogenas (IDH) 1 and IDH2 genes in control cerebellum and medulloblastoma samples, in a hotspot mutation (TIF 7812 KB)
Rights and permissions
About this article
Cite this article
Bezerra Salomão, K., Cruzeiro, G.A.V., Bonfim-Silva, R. et al. Reduced hydroxymethylation characterizes medulloblastoma while TET and IDH genes are differentially expressed within molecular subgroups. J Neurooncol 139, 33–42 (2018). https://doi.org/10.1007/s11060-018-2845-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-018-2845-1