Abstract
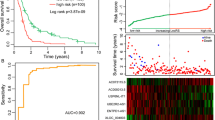
We investigated immune-related long non-coding RNAs (lncRNAs) that may be exploited as potential therapeutic targets in anaplastic gliomas. We obtained 572 lncRNAs and 317 immune genes from the Chinese Glioma Genome Atlas microarray and constructed immune-related lncRNAs co-expression networks to identify immune-related lncRNAs. Two additional datasets (GSE16011, REMBRANDT) were used for validation. Gene set enrichment analysis and principal component analysis were used for functional annotation. Immune-lncRNAs co-expression networks were constructed. Nine immune-related lncRNAs (SNHG8, PGM5-AS1, ST20-AS1, LINC00937, AGAP2-AS1, MIR155HG, TUG1, MAPKAPK5-AS1, and HCG18) signature was identified in patients with anaplastic gliomas. Patients in the low-risk group showed longer overall survival (OS) and progression-free survival than those in the high-risk group (P < 0.0001; P < 0.0001). Additionally, patients in the high-risk group displayed no-deletion of chromosomal arms 1p and/or 19q, isocitrate dehydrogenase wild-type, classical and mesenchymal TCGA subtype, G3 CGGA subtype, and lower Karnofsky performance score (KPS). Moreover, the signature was an independent factor and was significantly associated with the OS (P = 0.000, hazard ratio (HR) = 1.434). These findings were further validated in two additional datasets (GSE16011, REMBRANDT). Low-risk and high-risk groups displayed different immune status based on principal components analysis. Our results showed that the nine immune-related lncRNAs signature has prognostic value for anaplastic gliomas.




Similar content being viewed by others
Abbreviations
- AG:
-
Anaplastic glioma
- OS:
-
Overall survival
- lncRNA:
-
Long noncoding RNA
- CGGA:
-
Chinese Glioma Genome Atlas
- PCA:
-
Principal components analysis
- GSEA:
-
Gene set enrichment analysis
References
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114(2):97–109. https://doi.org/10.1007/s00401-007-0243-4
Yang P, Wang Y, Peng X, You G, Zhang W, Yan W, Bao Z, Wang Y, Qiu X, Jiang T (2013) Management and survival rates in patients with glioma in China (2004–2010): a retrospective study from a single-institution. J. Neuro-oncol 113(2):259–266. https://doi.org/10.1007/s11060-013-1103-9
Cheng W, Ren X, Zhang C, Cai J, Liu Y, Han S, Wu A (2016) Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology 86(24):2226–2234. https://doi.org/10.1212/WNL.0000000000002770
Silver DJ, Sinyuk M, Vogelbaum MA, Ahluwalia MS, Lathia JD (2016) The intersection of cancer, cancer stem cells, and the immune system: therapeutic opportunities. Neuro Oncol 18(2):153–159. https://doi.org/10.1093/neuonc/nov157
Finocchiaro G, Pellegatta S (2011) Immunotherapy for glioma: getting closer to the clinical arena? Curr Opin Neurol 24(6):641–647. https://doi.org/10.1097/WCO.0b013e32834cbb17
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10(3):155–159. https://doi.org/10.1038/nrg2521
Fang Y, Fullwood MJ (2016) Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinf. https://doi.org/10.1016/j.gpb.2015.09.006
Bian EB, Li J, Xie YS, Zong G, Li J, Zhao B (2015) LncRNAs: new players in gliomas, with special emphasis on the interaction of lncRNAs With EZH2. J Cell Physiol 230(3):496–503. https://doi.org/10.1002/jcp.24549
Sun Y, Wang Z, Zhou D (2013) Long non-coding RNAs as potential biomarkers and therapeutic targets for gliomas. Med Hypotheses 81(2):319–321. https://doi.org/10.1016/j.mehy.2013.04.010
Zhang XQ, Leung GK (2014) Long non-coding RNAs in glioma: functional roles and clinical perspectives. Neurochem Int 77:78–85. https://doi.org/10.1016/j.neuint.2014.05.008
Wang P, Ren Z, Sun P (2012) Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem 113(6):1868–1874. https://doi.org/10.1002/jcb.24055
Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA (2013) A long noncoding RNA mediates both activation and repression of immune response genes. Science 341(6147):789–792. https://doi.org/10.1126/science.1240925
Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, Braun JE, Gandhi P, Moore MJ, Chang HY, Lodish HF, Caffrey DR, Fitzgerald KA (2016) A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165(7):1672–1685. https://doi.org/10.1016/j.cell.2016.05.075
Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, Wang K, Jia W, Chu WM, Sun B (2017) The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun 8:15129. https://doi.org/10.1038/ncomms15129
Wang Q, Zhang J, Liu Y, Zhang W, Zhou J, Duan R, Pu P, Kang C, Han L (2016) A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett 373(2):251–259. https://doi.org/10.1016/j.canlet.2016.01.039
Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X, Sun T, Xie X, Zhou Y, Du Z (2016) Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis 7:e2123. https://doi.org/10.1038/cddis.2015.407
Zhang CB, Zhu P, Yang P, Cai JQ, Wang ZL, Li QB, Bao ZS, Zhang W, Jiang T (2015) Identification of high risk anaplastic gliomas by a diagnostic and prognostic signature derived from mRNA expression profiling. Oncotarget 6(34):36643–36651. https://doi.org/10.18632/oncotarget.5421
Cheng W, Ren X, Cai J, Zhang C, Li M, Wang K, Liu Y, Han S, Wu A (2015) A five-miRNA signature with prognostic and predictive value for MGMT promoter-methylated glioblastoma patients. Oncotarget 6(30):29285–29295. https://doi.org/10.18632/oncotarget.4978
Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK (2012) Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis 48(1):1–8. https://doi.org/10.1016/j.nbd.2012.06.004
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Cai J, Zhang W, Yang P, Wang Y, Li M, Zhang C, Wang Z, Hu H, Liu Y, Li Q, Wen J, Sun B, Wang X, Jiang T, Jiang C (2015) Identification of a 6-cytokine prognostic signature in patients with primary glioblastoma harboring M2 microglia/macrophage phenotype relevance. PLoS ONE 10(5):e0126022. https://doi.org/10.1371/journal.pone.0126022
Bao ZS, Li MY, Wang JY, Zhang CB, Wang HJ, Yan W, Liu YW, Zhang W, Chen L, Jiang T (2014) Prognostic value of a nine-gene signature in glioma patients based on mRNA expression profiling. CNS Neurosci Ther 20(2):112–118. https://doi.org/10.1111/cns.12171
Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R (2004) Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med 350(18):1828–1837. https://doi.org/10.1056/NEJMoa032520
Zhang W, Zhang J, Yan W, You G, Bao Z, Li S, Kang C, Jiang C, You Y, Zhang Y, Chen CC, Song SW, Jiang T (2013) Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer 119(4):814–824. https://doi.org/10.1002/cncr.27826
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Wang ZL, Zhang CB, Cai JQ, Li QB, Wang Z, Jiang T (2015) Integrated analysis of genome-wide DNA methylation, gene expression and protein expression profiles in molecular subtypes of WHO II-IV gliomas. J Exp Clin Cancer Res 34:127. https://doi.org/10.1186/s13046-015-0249-z
Wang W, Zhang L, Wang Z, Yang F, Wang H, Liang T, Wu F, Lan Q, Wang J, Zhao J (2016) A three-gene signature for prognosis in patients with MGMT promoter-methylated glioblastoma. Oncotarget. https://doi.org/10.18632/oncotarget.11726
Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y (2015) The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget 6(11):9160–9172. https://doi.org/10.18632/oncotarget.3247
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820. https://doi.org/10.1007/s00401-016-1545-1
Wirsching HG, Weller M (2016) The role of molecular diagnostics in the management of patients with gliomas. Curr Treat Options Oncol 17(10):51. https://doi.org/10.1007/s11864-016-0430-4
Bai H, Harmanci AS, Erson-Omay EZ, Li J, Coskun S, Simon M, Krischek B, Ozduman K, Omay SB, Sorensen EA, Turcan S, Bakirciglu M, Carrion-Grant G, Murray PB, Clark VE, Ercan-Sencicek AG, Knight J, Sencar L, Altinok S, Kaulen LD, Gulez B, Timmer M, Schramm J, Mishra-Gorur K, Henegariu O, Moliterno J, Louvi A, Chan TA, Tannheimer SL, Pamir MN, Vortmeyer AO, Bilguvar K, Yasuno K, Gunel M (2016) Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet 48(1):59–66. https://doi.org/10.1038/ng.3457
Hong H, Stastny M, Brown C, Chang WC, Ostberg JR, Forman SJ, Jensen MC (2014) Diverse solid tumors expressing a restricted epitope of L1-CAM can be targeted by chimeric antigen receptor redirected T lymphocytes. J Immunother 37(2):93–104. https://doi.org/10.1097/CJI.0000000000000018
Sampson JH, Choi BD, Sanchez-Perez L, Suryadevara CM, Snyder DJ, Flores CT, Schmittling RJ, Nair SK, Reap EA, Norberg PK, Herndon JE 2nd, Kuan CT, Morgan RA, Rosenberg SA, Johnson LA (2014) EGFRvIII mCAR-modified T-cell therapy cures mice with established intracerebral glioma and generates host immunity against tumor-antigen loss. Clin Cancer Res 20 (4):972–984. https://doi.org/10.1158/1078-0432.CCR-13-0709
Fine HA (2015) New strategies in glioblastoma: exploiting the new biology. Clin Cancer Res 21(9):1984–1988. https://doi.org/10.1158/1078-0432.CCR-14-1328
Cheng W, Li M, Jiang Y, Zhang C, Cai J, Wang K, Wu A (2016) Association between small heat shock protein B11 and the prognostic value of MGMT promoter methylation in patients with high-grade glioma. J Neurosurg 125(1):7–16. https://doi.org/10.3171/2015.5.JNS142437
Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, Kroeger P, McWeeny K, Halbert DN, Mollison KW, Djuric SW, Trevillyan JM (2002) T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol 217(1–2):78–86
van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, Poppema S (2003) High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosom Cancer 37(1):20–28. https://doi.org/10.1002/gcc.10186
Zhang T, Nie K, Tam W (2008) BIC is processed efficiently to microRNA-155 in Burkitt lymphoma cells. Leukemia 22(9):1795–1797. https://doi.org/10.1038/leu.2008.62
Wu X, Wang Y, Yu T, Nie E, Hu Q, Wu W, Zhi T, Jiang K, Wang X, Lu X, Li H, Liu N, Zhang J, You Y (2017) Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro Oncol. https://doi.org/10.1093/neuonc/nox017
Wang W, Zhang L, Wang Z, Yang F, Wang H, Liang T, Wu F, Lan Q, Wang J, Zhao J (2016) A three-gene signature for prognosis in patients with MGMT promoter-methylated glioblastoma. Oncotarget 7(43):69991–69999. https://doi.org/10.18632/oncotarget.11726
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106(28):11667–11672. https://doi.org/10.1073/pnas.0904715106
Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, Shibata T, Naito M, Kim HJ, Miyata K, Kataoka K, Kondo Y (2016) Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun 7:13616. https://doi.org/10.1038/ncomms13616
Wang W, Gao F, Zhao Z, Wang H, Zhang L, Zhang D, Zhang Y, Lan Q, Wang J, Zhao J (2017) Integrated analysis of lncRNA-mRNA co-expression profiles in patients with moyamoya disease. Sci Rep 7:42421. https://doi.org/10.1038/srep42421
Xu J, Zhang F, Gao C, Ma X, Peng X, Kong D, Hao J (2016) Microarray analysis of lncRNA and mRNA expression profiles in patients with neuromyelitis optica. Mol Neurobiol. https://doi.org/10.1007/s12035-016-9754-0
Li H, Wang W, Zhang L, Lan Q, Wang J, Cao Y, Zhao J (2017) Identification of a long noncoding RNA-associated competing endogenous RNA network in intracranial aneurysm. World Neurosurg 97:684–692. https://doi.org/10.1016/j.wneu.2016.10.016
Reon BJ, Anaya J, Zhang Y, Mandell J, Purow B, Abounader R, Dutta A (2016) Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: an in silico analysis. PLoS Med 13(12):e1002192. https://doi.org/10.1371/journal.pmed.1002192
Acknowledgements
We thank Dr. Hao Li for his assistance in analyzing the data.
Funding
This work was supported by grants from Ministry of Science and Technology of China Grant (2012CB825505 and 2011BAI08B08); National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2013BAI09B03, 2014BAI04B02); National High Technology Research and Development Program (No. 2012AA02A508); National Natural Science Foundation of China (No. 91229121); Beijing Municipal Administration of Hospitals’ Mission Plan (SML20150501); “13th Five-Year Plan” National Science and Technology supporting plan (2015BAI09B04).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11060_2017_2667_MOESM1_ESM.tif
Figure S1. Construction of immune-related lncRNAs co-expression networks. A. Immune-related lncRNAs co-expression networks based on 572 lncRNAs and 317 immune genes (P ≤ 0.001). B. Immune-related lncRNAs co-expression networks based on nine prognostic lncRNAs (P ≤ 0.001). Red, lncRNAs; Green, mRNAs. (TIF 5336 KB)
11060_2017_2667_MOESM2_ESM.tif
Figure S2. Kaplan-Meier curves for OS and PFS among AG patients from different groups stratified by the status of 1p and/or 19q and IDH in three datasets. (TIF 811 KB)
Rights and permissions
About this article
Cite this article
Wang, W., Zhao, Z., Yang, F. et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J Neurooncol 136, 263–271 (2018). https://doi.org/10.1007/s11060-017-2667-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-017-2667-6