Abstract
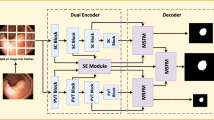
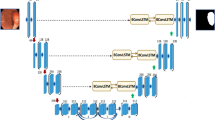
The incidence of colon cancer occupies the top three places in gastrointestinal tumors, and colon polyps are an important causative factor in the development of colon cancer. Early screening for colon polyps and colon polypectomy can reduce the chances of colon cancer. The current means of colon polyp examination is through colonoscopy, taking images of the gastrointestinal tract, and then manually marking them manually, which is time-consuming and labor-intensive for doctors. Therefore, relying on advanced deep learning technology to automatically identify colon polyps in the gastrointestinal tract of the patient and segmenting the polyps is an important direction of research nowadays. Due to the privacy of medical data and the non-interoperability of disease information, this paper proposes a dual-branch colon polyp segmentation network based on federated learning, which makes it possible to achieve a better training effect under the guarantee of data independence, and secondly, the dual-branch colon polyp segmentation network proposed in this paper adopts the two different structures of convolutional neural network (CNN) and Transformer to form a dual-branch structure, and through layer-by-layer fusion embedding, the advantages between different structures are realized. In this paper, we also propose the Aggregated Attention Module (AAM) to preserve the high-dimensional semantic information and to complement the missing information in the lower layers. Ultimately our approach achieves state of the art in Kvasir-SEG and CVC-ClinicDB datasets.










Similar content being viewed by others
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
McMahan B, Moore E, Ramage D, Hampson S, y Arcas BA (2017) Communication-efficient learning of deep networks from decentralized data. In: Artificial intelligence and statistics, pp 1273–1282. PMLR
Li T, Sahu AK, Zaheer M, Sanjabi M, Talwalkar A, Smith V (2018) Federated optimization in heterogeneous networks
Li X, Jiang M, Zhang X, Kamp M, Dou Q (2021) FedBN: federated learning on non-iid features via local batch normalization
Li Q, He B, Song D (2021) Model-contrastive federated learning. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 10713–10722
Wang J, Liu Q, Liang H, Joshi G, Poor HV (2020) Tackling the objective inconsistency problem in heterogeneous federated optimization
Long J, Shelhamer E, Darrell T (2014) Fully convolutional networks for semantic segmentation
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. Springer, pp 234–241
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2018) UNet++: a nested u-net architecture for medical image segmentation
Diakogiannis FI, Waldner F, Caccetta P, Wu C (2020) ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data. ISPRS J Photogrammetr Remote Sens 162:94–114
Badrinarayanan V, Kendall A, Cipolla R (2015) SegNet: a deep convolutional encoder-decoder architecture for image segmentation
Carion N, Massa F, Synnaeve G, Usunier N, Kirillov A, Zagoruyko S (2020) End-to-end object detection with transformers
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, Uszkoreit J, Houlsby N (2020) An image is worth 16x16 words: transformers for image recognition at scale
Wang W, Xie E, Li X, Fan D-P, Song K, Liang D, Lu T, Luo P, Shao L (2021) PVT v2: improved baselines with pyramid vision transformer. https://doi.org/10.1007/s41095-022-0274-8
Wang W, Xie E, Li X, Fan D-P, Song K, Liang D, Lu T, Luo P, Shao L (2021) Pyramid vision transformer: a versatile backbone for dense prediction without convolutions
Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B (2021) Swin transformer: hierarchical vision transformer using shifted windows
Liu Z, Hu H, Lin Y, Yao Z, Xie Z, Wei Y, Ning J, Cao Y, Zhang Z, Dong L, Wei F, Guo B (2021) Swin transformer v2: scaling up capacity and resolution
Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S (2016) Feature pyramid networks for object detection
He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition
Zhao H, Shi J, Qi X, Wang X, Jia J (2016) Pyramid scene parsing network
Jha D, Smedsrud PH, Riegler MA, Halvorsen P, Johansen HD (2020) Kvasir-SEG: a segmented polyp dataset
Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F (2015) WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput Med Imag Graph 43:99–111
Jiang Y, Konečný J, Rush K, Kannan S (2019) Improving federated learning personalization via model agnostic meta learning
Sater RA, Hamza AB (2021) A federated learning approach to anomaly detection in smart buildings
Verma R, Kumar N, Patil A, Kurian NC, Rane S, Graham S, Vu QD, Zwager M, Raza SEA, Rajpoot N, Wu X, Chen H, Huang Y, Wang L, Jung H, Brown GT, Liu Y, Liu S, Jahromi SAF, Khani AA, Montahaei E, Baghshah MS, Behroozi H, Semkin P, Rassadin A, Dutande P, Lodaya R, Baid U, Baheti B, Talbar S, Mahbod A, Ecker R, Ellinger I, Luo Z, Dong B, Xu Z, Yao Y, Lv S, Feng M, Xu K, Zunair H, Hamza AB, Smiley S, Yin TK, Fang QR, Srivastava S, Mahapatra D, Trnavska L, Zhang H, Narayanan PL, Law J, Yuan Y, Tejomay A, Mitkari A, Koka D, Ramachandra V, Kini L, Sethi A (2021) MoNuSAC 2020: a multi-organ nuclei segmentation and classification challenge. IEEE Trans Med Imag 40:3413–3423. https://doi.org/10.1109/TMI.2021.3085712
Zunair H, Hamza AB (2022) Masked supervised learning for semantic segmentation
Hou Q, Zhang L, Cheng M-M, Feng J (2020) Strip pooling: rethinking spatial pooling for scene parsing
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I (2017) Attention is all you need
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) ImageNet: a large-scale hierarchical image database. IEEE, pp 248–255
Wu Y, He K (2018) Group normalization
Elfwing S, Uchibe E, Doya K (2018) Sigmoid-weighted linear units for neural network function approximation in reinforcement learning. Neural Netw 107:3–11
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift
Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. In: Proceedings of the fourteenth international conference on artificial intelligence and statistics, pp 315–323. JMLR Workshop and Conference Proceedings
Wang X, Girshick R, Gupta A, He K (2018) Non-local neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7794–7803
Fan D-P, Ji G-P, Zhou T, Chen G, Fu H, Shen J, Shao L (2020) PraNet: parallel reverse attention network for polyp segmentation
Wei J, Wang S, Huang Q (2020) F\(^3\)net: fusion, feedback and focus for salient object detection, vol 34, pp 12321–12328
Loshchilov I, Hutter F (2017) Decoupled weight decay regularization
Fitzgerald K, Matuszewski B (2023) Fcb-swinv2 transformer for polyp segmentation. arXiv preprint arXiv:2302.01027
Sanderson E, Matuszewski BJ (2022) FCN-transformer feature fusion for polyp segmentation, vol 13413 LNCS. Springer, pp 892–907. https://doi.org/10.1007/978-3-031-12053-4_65
Liao T-Y, Yang C-H, Lo Y-W, Lai K-Y, Shen P-H, Lin Y-L (2022) HarDNet-DFUS: an enhanced harmonically-connected network for diabetic foot ulcer image segmentation and colonoscopy polyp segmentation
Rahman MM, Marculescu R (2023) Medical image segmentation via cascaded attention decoding. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp 6222–6231
Funding
This research was funded by National Natural Science Foundation of China (General Program, (No.62271456), and National Key Science and Technology Program 2030 (No. 2021ZD0110600).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, X., Fan, K. & Ma, H. Federal learning-based a dual-branch deep learning model for colon polyp segmentation. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-19197-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-19197-6