Abstract
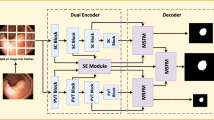
Colorectal Cancer (CRC) is one of the most common cancer diseases in the world. Early diagnosis of the disease is of great importance for the recovery of the patient. Colonoscopy is the gold standard procedure used in the diagnosis of CRC. In this context, this study focused on the detection of polyps with high accuracy in order to contribute to the early diagnosis of CRC. Within the scope of the study, polyp segmentation was performed on the public CVC-Clinic DB polyp dataset. In the study, the basic U-Net model and its derivatives (modified U-Net, modified U-Net with transfer learning (VGG-16, VGG-19) in the encoding part) were used for the segmentation process. For sensitivity analysis, models were trained on three separate datasets prepared with different preprocessing methods in addition to the raw dataset with k-fold cross validations (k = 2,3,4) and different batch numbers (1,2,3,4,5) in each cross validation. As a result of the analysis, the best performance was obtained as 0.868, 0.799, 0.873 and 0.994 for Dice, Jaccard, Sensitivity, Specificity when the batch size was taken as 1 with fourfold cross validation in the modified U-Net trained with the Discrete Wavelet Transform (DWT) dataset. This model and its parameters were then tested with public datasets Kvasir-Seg and Etis-Larib Polyp DB. Moreover, different models were trained with the parameters of the most successful model. The results of all analyzes were interpreted and compared with the literature.












Similar content being viewed by others
Data availability
Publicly available datasets were analyzed in this study.
The CVC-Clinic DB datasets are publicly available here: https://polyp.grand-challenge.org/CVCClinicDB/ (accessed on 10 August 2022).
The ETIS-Larib dataset is publicly available here: https://polyp.grand-challenge.org/EtisLarib (accessed on 10 August 2022).
The Kvasir-SEG dataset is publicly available here: https://datasets.simula.no/kvasir-seg/ (accessed on 10 August 2022).
References
Abraham N, Khan NM A novel focal tversky loss function with improved attention u-net for lesion segmentation. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), 2019. IEEE, pp 683–687
Ahmed N, Natarajan T, Rao KR (1974) Discrete Cosine Transform. IEEE Trans Comput C–23(1):90–93. https://doi.org/10.1109/T-C.1974.223784
Arnold M, Ghosh A, Ameling S, Lacey G (2010) Automatic segmentation and inpainting of specular highlights for endoscopic imaging. EURASIP J Image Video Process 2010:1–12
Ashkani Chenarlogh V, Shabanzadeh A, Ghelich Oghli M, Sirjani N, Farzin Moghadam S, Akhavan A, Arabi H, Shiri I, Shabanzadeh Z, Sanei Taheri M (2022) Clinical target segmentation using a novel deep neural network: double attention Res-U-Net. Sci Rep 12(1):6717
Banik D, Bhattacharjee D, Nasipuri M (2020) A multi-scale patch-based deep learning system for polyp segmentation. In: Advanced Computing and Systems for Security. Springer 12:109–119
Banik D, Roy K, Bhattacharjee D, Nasipuri M, Krejcar O (2020) Polyp-Net: A multimodel fusion network for polyp segmentation. IEEE Trans Instrum Meas 70:1–12
Bardhi O, Sierra-Sosa D, Garcia-Zapirain B, Elmaghraby A (2017) Automatic colon polyp detection using Convolutional encoder-decoder model. In: IEEE international symposium on signal processing and information technology (ISSPIT). IEEE, pp 445–448. https://doi.org/10.1109/ISSPIT.2017.8388684
Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F (2015) WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput Med Imaging Graph 43:99–111. https://doi.org/10.1016/j.compmedimag.2015.02.007
Bernal J, Sánchez J, Vilarino F (2012) Towards automatic polyp detection with a polyp appearance model. Pattern Recogn 45(9):3166–3182
Bernal J, Tajkbaksh N, Sánchez FJ, Matuszewski BJ, Chen H, Yu L, Angermann Q, Romain O, Rustad B, Balasingham I (2017) Comparative validation of polyp detection methods in video colonoscopy: results from the MICCAI 2015 endoscopic vision challenge. IEEE Trans Med Imaging 36(6):1231–1249
Brandao P, Mazomenos E, Ciuti G, Caliò R, Bianchi F, Menciassi A, Dario P, Koulaouzidis A, Arezzo A, Stoyanov D (2017) Fully convolutional neural networks for polyp segmentation in colonoscopy. In: Medical Imaging: Computer-Aided Diagnosis. Spie 10134:101–107
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. International conference on medical image computing and computer-assisted intervention. Springer, pp 424–432
Hu Y, Li Q, Ma S, Kuo C-C A VLSI architecture for a fast computation of the 2-D discrete wavelet transform. In: 2007 IEEE International Symposium on Circuits and Systems (ISCAS), 2007. IEEE, pp 3980–3983
Huttenlocher DP, Klanderman GA, Rucklidge WJ (1993) Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Mach Intell 15(9):850–863
Jha D, Smedsrud PH, Riegler MA, Halvorsen P, De Lange T, Johansen D, Johansen HD (2020) Kvasir-seg: A segmented polyp dataset. In: International Conference on Multimedia Modeling, Springer, pp 451-4625
Kang J, Gwak J (2019) Ensemble of instance segmentation models for polyp segmentation in colonoscopy images. IEEE Access 7:26440–26447
Kolligs FT (2016) Diagnostics and Epidemiology of Colorectal Cancer. Visc Med 32(3):158–164. https://doi.org/10.1159/000446488
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems 25
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Li Q, Yang G, Chen Z, Huang B, Chen L, Xu D, Zhou X, Zhong S, Zhang H, Wang T (2017) Colorectal polyp segmentation using a fully convolutional neural network. In: 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), 2017: 1–5. https://doi.org/10.1109/CISP-BMEI.2017.8301980
Lin T-Y, Goyal P, Girshick R, He K, Dollár P Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision, 2017. pp 2980–2988
Nguyen Q, Lee S-W Colorectal segmentation using multiple encoder-decoder network in colonoscopy images. In: 2018 IEEE first international conference on artificial intelligence and knowledge engineering (AIKE), 2018. IEEE, pp 208–211
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B (2018) Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:180403999
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Schmeelk J (2002) Wavelet transforms on two-dimensional images. Math Comput Model 36(7):939–948. https://doi.org/10.1016/S0895-7177(02)00238-8
Shorten C, Khoshgoftaar TM (2019) A survey on Image Data Augmentation for Deep Learning. J Big Data 6(1):60. https://doi.org/10.1186/s40537-019-0197-0
Silva J, Histace A, Romain O, Dray X, Granado B (2014) Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. Int J Comput Assist Radiol Surg 9(2):283–293. https://doi.org/10.1007/s11548-013-0926-3
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556
Society AC (2021) Cancer Facts & Figures 2021. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2021/cancer-facts-and-figures-2021.pdf. Accessed 30 Aug 2022
Stone M (1974) Cross-validatory choice and assessment of statistical predictions. J Roy Stat Soc: Ser B (Methodol) 36(2):111–133
Williams CB (2009) Insertion Technique. In: Colonoscopy. pp 535–559. https://doi.org/10.1002/9781444316902.ch40
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2018) Unet++: A nested u-net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop Proceedings 4. Springer pp 3–11
Zuiderveld K (1994) VIII.5. - Contrast Limited Adaptive Histogram Equalization. In: Heckbert PS (ed) Graphics Gems. Academic Press, pp 474–485. https://doi.org/10.1016/B978-0-12-336156-1.50061-6
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Solak, A., Ceylan, R. A sensitivity analysis for polyp segmentation with U-Net. Multimed Tools Appl 82, 34199–34227 (2023). https://doi.org/10.1007/s11042-023-16368-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-16368-9