Abstract
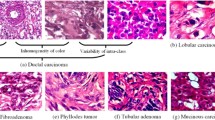
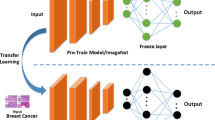
On the global level, Invasive Ductal Carcinoma (IDC) is one of the leading malignancies among women undergoing breast cancer screening. A slight delay in detecting and diagnosing this disease may result in irreversible complications. Histopathological images obtained from a biopsy examination present enormous structural information, which helps significantly in improving the prognosis of the disease. Pathological analysis involving microscopic examination of the histopathological slides is a highly challenging task due to the limited abilities of conventional computer-aided detection (CAD) methods to reach a precise diagnosis. The problem has become more accentuated due to the less availability of medical data in the public domain. Lately, deep learning methods using artificial neural networks are being used consistently to improve the performance of these CAD methods. Despite less medical data availability, transfer learning has recently been commonly practised to facilitate a deep neural network to train on a specific dataset in resolving the problem. However, the performance of this method is not remarkably appreciable on small and low-resolution medical image datasets compared to those comprising whole slide images of higher resolutions. In this direction, this study proposes a domain-specific learning strategy, Histo-Fusion, with the objective of detecting the IDC more precisely. In the proposed method, the deep CNN model is initially trained on higher-resolution histopathological images of breast tissue which presents ample information for a model to learn the significant features for better discrimination between normal and malignant tissues. Subsequently, through a positive transfer of domain features, the model is further trained on the small and low-resolution images, enabling it to classify these histology images into IDC - and IDC + categories. Moreover, shallow and deep neural network architectures are utilized in the study to compare their performance across the two learning approaches: transfer learning and Histo-Fusion on the IDC dataset. As revealed by the present study results, the proposed Histo-Fusion learning approach has improved each deep CNN’s discriminating abilities by yielding better accuracy scores of around 5% over and above those obtained by the commonly used transfer learning strategy. Therefore, the procedure is expected to reduce false-positive rates and help expert pathologists reach accurate diagnoses.







Similar content being viewed by others
Data availability
The data used in the study is freely available and accessible to research community on following links:
1. https://www.kaggle.com/paultimothymooney/breast-histopathology-images
2. https://www.kaggle.com/datasets/ambarish/breakhis/download
References
Alanazi SA, Kamruzzaman MM, Islam Sarker MN, Alruwaili M, Alhwaiti Y, Alshammari N, Siddiqi MH (2021) Boosting breast cancer detection using convolutional neural network. J Healthc Eng 2021
Alghodhaifi H, Alghodhaifi A, Alghodhaifi M (2019) Predicting invasive ductal carcinoma in breast histology images using convolutional neural network. In: 2019 IEEE National Aerospace and Electronics Conference (NAECON). IEEE, pp 374–378
Alghodhaifi H, Alghodhaifi A, Alghodhaifi M (2019) Predicting invasive ductal carcinoma in breast histology images using convolutional neural network. In: Proceedings of the 2019 IEEE National Aerospace and electronics conference (NAECON), Dayton, pp 15–19
Alzubaidi L, Al-Amidie M, Al-Asadi A, Humaidi AJ, Al-Shamma O, Fadhel MA, Duan Y (2021) Novel transfer learning approach for medical imaging with limited labeled data. Cancers 13(7):1590
Attallah O (2019) Multi-tasks biometric system for personal identification. In: IEEE international conference on Computational Science and Engineering (CSE) and IEEE international conference on Embedded and Ubiquitous Computing (EUC). IEEE, pp 110–114
Attallah O, Anwar F, Ghanem NM, Ismail MA (2021) Histo-CADx: duo cascaded fusion stages for breast cancer diagnosis from histopathological images. PeerJ Computer Science 7:e493
Barsha NA, Rahman A, Mahdy MRC (2021) Automated detection and grading of invasive ductal carcinoma breast cancer using ensemble of deep learning models. Comput Biol Med 139:104931
Bengio Y (2009) Learning deep architectures for AI. Found Trends Mach Learn, pp 1–127
Boser BE, Guyon IM (1992) A training algorithm for optimal margin classifiers. In: Fifth annual workshop on Computational Learning Theory, ACM, pp 144–152
Breiman L (2001) Random forests Machine learning 45:5–32
Chan A, Tuszynski JA (2016) Automatic prediction of tumour malignancy in breast cancer with fractal dimension. R Soc Open Sci 3(12):160558
Chapala H, Sujatha B (2020) ResNet: detection of invasive ductal carcinoma in breast histopathology images using deep learning. In: 2020 international conference on electronics and sustainable communication systems (ICESC). IEEE, pp 60–67
Chen J (2021) A transfer learning based super-resolution microscopy for biopsy slice images: the joint methods perspective. IEEE/ACM Trans Comput Biol Bioinf, pp 103–113
Chen Z, Gao R, Mao K, Wang P, Yan R, Zhao R (2016) Deep learning and its applications to machine health monitoring: a survey. CoRR:abs/1612.07640
Coelho LP, Ahmed A, Arnold A, Kangas J, Sheikh AS, Xing EP, Murphy RF (2010) Structured literature image finder: extracting information from text and images in biomedical literature. In: Linking literature, information, and knowledge for biology: workshop of the BioLink special interest group, ISMB/ECCB 2009, Stockholm, June 28–29, 2009, revised selected papers. Springer, Berlin Heidelberg, pp 23–32
Cruz-Roa A, Basavanhally A, González F, Gilmore H, Feldman M, Ganesan S, Madabhushi A (2014) Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In: Medical imaging 2014: Digital Pathology, vol 9041. SPIE, p 904103
Elmore JG, Jackson SL, Abraham L, Miglioretti DL, Carney PA, Geller BM, Buist DS (2009) Variability in interpretive performance at screening mammography and radiologists’ characteristics associated with accuracy. Radiology 253(3):641–651
Feig SA (2014) Screening mammography benefit controversies: sorting the evidence. Radiol Clin 52(3):455–480
Gao H, Xiao J, Yin Y, Liu T, Shi J (2022) A mutually supervised graph attention network for few-shot segmentation: the perspective of fully utilizing limited samples. IEEE Trans neural Net Learn Syst
Gao H, Xu K, Cao M, Xiao J, Xu Q, Yin Y (2021) The deep features and attention mechanism-based method to dish healthcare under social iot systems: an empirical study with a hand-deep local–global net. IEEE Trans Comput Soc Syst 9(1):336–347
Hamidinekoo A, Denton E, Rampun A, Honnor K, Zwiggelaar R (2018) Deep learning in mammography and breast histology, an overview and future trends. Med Image Anal 47:45–67
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hedjazi MA, Kourbane I, Genc Y (2017) On identifying leaves: a comparison of CNN with classical ML methods. In: 2017 25th signal processing and communications applications conference (SIU). IEEE, pp 1–4
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Janowczyk A, Madabhushi A (2016) Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform 7(1):29
Jawad MA, Khursheed F (2022) Deep and dense convolutional neural network for multi category classification of magnification specific and magnification independent breast cancer histopathological images. Biomed Signal Process Control 78:103935
Kowal M, Filipczuk P, Obuchowicz A, Korbicz J, Monczak R (2013) Computer-aided diagnosis of breast cancer based on fine needle biopsy microscopic images. Comput Biol Med 43(10):1563–1572
Krizhevsky A, Sutskever I, Hinton GE (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Lehman CD, Wellman RD, Buist DS, Kerlikowske K, Tosteson AN, Miglioretti DL, & Breast Cancer Surveillance Consortium (2015) Diagnostic accuracy of digital screening mammography with and without computer-aided detection. JAMA Intern Med 175(11):1828–1837
Lepetit V, Fua P (2006) Keypoint recognition using randomized trees. IEEE Trans Pattern Anal Mach Intell 28(9):1465–1479
Li Y, Chen H, Cao L, Ma J (2016) A survey of computer-aided detection of breast cancer with mammography. J Health Med Inf 4(7):1–6
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Guan X, Thomas NE (2009, June) A method for normalizing histology slides for quantitative analysis. In: 2009 IEEE international symposium on biomedical imaging: from nano to macro. IEEE, pp 1107–1110
Mathur P, Sathishkumar K, Chaturvedi M, Das P, Sudarshan KL, Santhappan S, ICMR-NCDIR-NCRP Investigator Group (2020) Cancer statistics, 2020: report from national cancer registry programme, India. JCO Glob Oncol 6:1063–1075
Nanni L, Ghidoni S, Brahnam S (2017) Handcrafted vs. non-handcrafted features for computer vision classification. Pattern Recogn 71:158–172
Narayanan BN, Krishnaraja V, Ali R (2019) Convolutional neural network for classification of histopathology images for breast Cancer detection. IEEE National Aerospace and Electronics Conference (NAECON) 2019:291–295
Neal LT (2010) Clinician’s guide to imaging and pathologic findings in benign breast disease. In: Mayo Clinic proceedings, pp 274–279
Ojala T, Pietikainen M, Maenpaa T (2002) Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 24(7):971–987
Ojansivu V, Heikkila J (2008) Blur insensitive texture classification using local phase quantization. Lect Notes Comput Sci 5099:236–243
Prokhorenkova L, Gusev G, Vorobev A, Dorogush AV, Gulin A (2018) CatBoost: unbiased boosting with categorical features. Adv Neural Inf Proces Syst 31
Raghu M, Zhang C, Kleinberg J, Bengio S (2019) Transfusion: understanding transfer learning for medical imaging. Adv Neural Inf Proces Syst 32
Rahman MJU, Sultan RI, Mahmud F, Al Ahsan S, Matin A (2018) Automatic system for detecting invasive ductal carcinoma using convolutional neural networks. In: TENCON 2018–2018 IEEE region 10 conference. IEEE, pp 0673–0678
Robboy SJ, Weintraub S, Horvath AE, Jensen BW, Alexander CB, Fody EP, Black-Schaffer WS (2013) Pathologist workforce in the United States: I. development of a predictive model to examine factors influencing supply. Arch Pathol Lab Med 137(12):1723–1732
Romano AM, Hernandez AA (2019) Enhanced deep learning approach for predicting invasive ductal carcinoma from histopathology images. In: 2019 2nd International Conference on Artificial Intelligence and Big Data (ICAIBD). IEEE, pp 142–148
Romero FP, Tang A, Kadoury S (2019) Multi-level batch normalization in deep networks for invasive ductal carcinoma cell discrimination in histopathology images. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019). IEEE, pp 1092–1095
Roy SD, Das S, Kar D, Schwenker F, Sarkar R (2021) Computer aided breast cancer detection using ensembling of texture and statistical image features. Sensors 21(11):3628
Rublee E, Rabaud V, Konolige K, Bradski G (2011) ORB: an efficient alternative to SIFT or SURF. In: 2011 international conference on computer vision. IEEE, pp 2564–2571
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Fei-Fei L (2015) Imagenet large scale visual recognition challenge. Int J Comput Vis 115:211–252
Sharma S, Mehra R (2020) Conventional machine learning and deep learning approach for multi-classification of breast cancer histopathology images—a comparative insight. J Digit Imaging 33:632–654
Spanhol FA, Oliveira LS, Petitjean C, Heutte L (2015) A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng 63(7):1455–1462
Spanhol FA, Oliveira LS (2016) A dataset for breast Cancer histopathological image classification. IEEE Trans Biomed Eng:1455–1462
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Tangudu, N (2021) Computer aided diagnosis of breast cancer from histopathological images using deep learning techniques. Turk J Physiother Rehab
Tharwat A (2016) Linear vs. quadratic discriminant analysis classifier: a tutorial. International journal of applied. Pattern Recogn 3(2):145–180
Veta M, Pluim JP, Van Diest PJ, Viergever MA (2014) Breast cancer histopathology image analysis: a review. IEEE Trans Biomed Eng 61(5):1400–1411
Wang JL, Ibrahim AK, Zhuang H, Ali AM, Li AY, Wu A (2018) A study on automatic detection of IDC breast cancer with convolutional neural networks. In: 2018 international conference on computational science and computational intelligence (CSCI). IEEE, pp 703–708
Weinberger KQ, Saul LK (2009) Distance metric learning for large margin nearest neighbor classification. J Mach Learn Res 10(2)
Welch HG, Passow HJ (2014) Quantifying the benefits and harms of screening mammography. JAMA Intern Med 174(3):448–454
Xie J, Liu R, Luttrell J IV, Zhang C (2019) Deep learning based analysis of histopathological images of breast cancer. Front Genet 10:80
Zhang W, Deng L, Zhang L, Wu D (2022) A survey on negative transfer. IEEE/CAA J Autom Sin
Zhang X, Zhang Y, Qian B, Liu X, Li X, Wang X, Wang L (2019) Classifying breast cancer histopathological images using a robust artificial neural network architecture. In: Bioinformatics and biomedical engineering: 7th International Work-Conference, IWBBIO 2019, Granada, Spain, May 8–10, 2019, proceedings, part I 7. Springer International Publishing, pp 204–215
Funding
The authors received no funding for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that there are no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jawad, M.A., Khursheed, F. Histo-fusion: a novel domain specific learning to identify invasive ductal carcinoma (IDC) from histopathological images. Multimed Tools Appl 82, 39371–39392 (2023). https://doi.org/10.1007/s11042-023-15134-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15134-1