Abstract
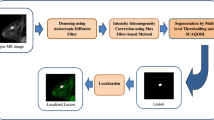
In recent years cancer on breast in women has increased rapidly worldwide. Therefore, the automatic segmentation of breast Dynamic Contrast-Enhanced Magnetic Resonance Imaging (DCE-MRI) has been exhaustively investigated since the proper use of methods permits the diagnosis and identification of diseases. Radiologists accept that breast fat-suppressed DCE-MRI evaluation for lesion detection and segmentation, which optimization algorithm via multi-level thresholding, is essential to differentiate breast lesions from other tissue types in DCE-MRI. This article proposes a breast DCE-MRI segmentation method using a multilevel thresholding technique based on enhanced Slime Mould Algorithm (SMA). The anisotropic diffusion filter is used to denoise MR images first. The preprocessing step then corrects intensity inhomogeneities. The suggested SMAQOBL algorithm is used to segment preprocessed MR images. Next, we developed the enhanced SMA by incorporating the Quasi Opposition-based Learning (QOBL) mechanism in it. This algorithm is used to find optimal threshold values through the maximization of Shannon entropy. Throughout this article, the proposed algorithm has been termed SMAQOBL. Finally, the segmented lesions are accurately localized in MR images. The proposed method is evaluated using 200 sagittal T2-weighted fat-suppressed DCE-MRI images of 40 patients. The SMAQOBL is compared with SMA, Dragonfly Optimization (DA), Grasshopper Optimization Algorithm (GOA), Particle Swarm Optimizer (PSO), Multi-Verse Optimization (MVO), Conventional Markov Random Field (CMRF), Hidden Markov Random Field (HMRF), and Improved Markov Random Field (IMRF). The best-achieved results of the proposed method in terms of accuracy is 99.94%, sensitivity is 99.86% and Dice Similarity Coefficient (DSC) is 98.41%. Evaluating the proposed method achieves an mean accuracy of 99.36%, a mean sensitivity of 95.83%, and mean DSC of 92.19%. We have analyzed the results using a one-way ANOVA test with posthoc Tukey-HSD test and Wilcoxon Signed Rank Test with Bonferroni correction. Furthermore, we have also analyzed the overall performance using Multi-Criteria Decision Making based on sensitivity, accuracy, specificity, Geometric-Mean, F-measure, DSC, and False Positive Rate (FPR). The proposed methods outperform other compared methods, according to both quantitative and qualitative outcomes.

























Similar content being viewed by others
Availability of data and material
Not Applicable.
Code availability
Not Applicable.
References
Agner SC, Xu J, Rosen M, Karthigeyan S, Englander S, Madabhushi A (2011) Spectral embedding based active contour: application to breast lesion segmentation on dce-mri. Proc. SPIE 7963, Medical Imaging 2011: Computer-Aided Diagnosis (796305), pp 280–290
Agliozzo S, Luca MD, Bracco C, Vignati A, Giannini V, Martincich L, Carbonaro LA, Bert A, Sardanelli F, Regge D (2012) Computer-aided diagnosis for dynamic contrast enhances breast mri of mass-like lesions using a multiparametric model combining a selection of morphological, kinetic, and spatiotemporal features. Med Phys 34:1704–1715
AlQoud A, Jaffar M A (2016) Hybrid gabor based local binary patterns texture features for classification of breast mammograms. International Journal of Computer Science and Network Security 16(4):16–21
Anscombe F (1948) The Validity of comparative experiments. J R Stat Soc 111(3):181–211
Arbach L, Stolpen A, Reinhardt JM (2004) Classification of Breast MRI Lesions using a Backpropagation Neural Network. In 2004 2nd IEEE international symposium on biomedical imaging: Macro to Nano (IEEE Cat No.04EX821 2:253–256
Arjmand A, Meshgini S, Afrouzian R, Farzamnia A (2019) Breast Tumor Segmentation Using K-Means Clustering and Cuckoo Search Optimization. In: 2019 9th International conference on computer and knowledge engineering (ICCKE) 305-308. https://doi.org/10.1109/iccke48569.2019.8964794
Azmi R, Norozi N (2011) A new markov random field segmentation method for breast lesion segmentation in mr images. Journal of Medical Signals Sensors 1:156–164
Balafar MA, Ramli AR, Mashohor S (2010) A new method for mr grayscale inhomogeneity correction. Artif Intell Rev 34:195–204
Behrens S, Laue H, Althaus M, Boehler T, Kuemmerlen B, Hahn HK, Peitgen HO (2007) Computer assistance for mr based diagnosis of breast cancer: present and future challenges. Comput Med Imaging Graph 31:236–247
Benjelloun M, Adoui ME, Larhmam MA, Mahmoudi SA (2018) Auto-mated breast tumor segmentation in DCE-MRI using deep learning. In: 4th International Conference on Cloud Computing Technologies and Applications (Cloudtech). https://doi.org/10.1109/CloudTech.2018.8713352
Bergstra J, Bengio Y (2012) Random search for hyper-parameter optimization. J Mach Learn Res 13:281–305
Bohare MD, Cheeran AN, Sarode VG (2011) Analysis of breast mri images using wavelets for detection of cancer. IJCA Special Issue on Electronics. Inf Commun Eng 4:1–3
Boukerroui D, Basset O, Guerin N, Baskurt A (1998) Multiresolution texture based adaptive clustering algorithm for breast lesion segmentation. Eur J Ultrasound 8:135–144
Bray F, Ren JS, Masuyer E (2013) Estimates of global cancer prevalence for 27 sites in the adult population in 2008. International Journal Cancer 132(5):1133–1145
Brown S, Tauler R, Walczak B (2020) Comprehensive Chemometrics- Chemical and Biochemical Data Analysis. 2nd Edition
Chatzis SP, Tsechpenakis G (2010) The infinite hidden markov random field model. IEEE Transactions On Neural Networks 21(6):1004–1014
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F (2013) The cancer imaging archive: Maintaining and operating a public information repository. Jounal Digit Imaging 26(6):1045–1057
Clerc M (1999) The swarm and the queen: Towards a deterministic and adaptive particle swarm optimization. In: Proceedings of the 1999 congress on evolutionary computation-CEC99 I: pp 1951–1957
Cross GR, Jain AK (1983) Markov random field texture models. IEEE Transactions On Pattern Analysis And Machine Intelligence 5(1):25–39
Derrac J, Garcia S, Molina D (2001) A practical tutorial on the use of nonparametric statistical tests as a methodology for comparing evolutionary and swarm intelligence algorithms. Swarm and Evolutionary Springer Nature, 2021. LATEX template 40 Breast DCE-MRI segmentation using SMAQOBL Computation 1:3–18
Dhane DM, Maity M, Achar A, Bar C, Chakraborty C (2015) Selection of optimal denoising filter using quality assessment for potentially lethal optical wound images. Procedia Comput Sci 58:438–446
Eltoukhy MM, Faye I, Samir BB (2010) Breast cancer diagnosis in digital mammogram using multiscale curvelet transform. Comput Med Imaging Graph 34(4):269–276
Engelbrecht AP (2005) Fundamentals of Computational Swarm Intelligence, John Wiley Sons Ltd
Eskandari P, Baradaran Shokouhi S (2021) Automated atlas-based segmentation of breast region in 3-D magnetic resonance imaging (MRI) using FCM method. Mapta Journal of Electrical and Computer Engineering Springer Nature 2021 LATEX template 42 Breast DCE-MRI segmentation using SMAQOBL (MJECE) 3(1):28–34. https://doi.org/10.33544/mjece.v3i1.175
Ferlay J, Soerjomataram I, Ervik M (2013) Globocan 2012 v1.0, cancer incidence and mortality worldwide: Iarc cancer base. GLOBOCAN
Ha W, Vahedi Z (2021) Automatic breast tumor diagnosis in mri based on a hybrid cnn and feature-based method using improved deer hunting opti- mization algorithm. Comput Intell Neurosci 2021(3):1–11. https://doi.org/10.1155/2021/5396327
Hauth EA, Stockamp C, Maderwald S, Muhler A, Kimmig R, Jaeger H, Barkhausen J, Forsting M (2006) Evaluation of the three-time-point method for diagnosis of breast lesions in contrast-enhanced mr mammography. Clinic Imaging 30:160–165
Huynh-Thu Q, Ghanbari M (2008) Scope of validity of PSNR in image/video quality assessment. Electron Lett 44:800–801
Hommel G (1988) A stagewise rejective multiple test procedure based on a modified bonferroni test. Biometroka 75(2):383–386
Jiao Z, Gao X, Wang Y, Li J (2016) A deep feature based framework for breast masses classification. Computer-Aided Diagnosis in Medical Imaging 197:221–231
Kadry S, Damasevicius R, Taniar D, Rajinikanth V, Lawal I A (2021) Extraction of Tumour in Breast MRI using Joint Thresholding and Seg- mentation – A Study, Seventh International conference on Bio Signals, Images, and Instrumentation (ICBSII). https://doi.org/10.1109/ICBSII51839.2021.9445152
Karthiga R, Narasimhan K (2018) Automated diagnosis of breast cancer using wavelet based entropy features. Second international conference on electronics, Communication and Aerospace Technology, pp 274–279. https://doi.org/10.1109/ICECA.2018.8474739
Kashyap KL, Bajpai MK, Khanna P (2015) Breast cancer detection in digital mammograms. In: 2015 IEEE international conference on imaging systems and techniques (IST) pp 1-6
Keyvanfard F, Shoorehdeli MA, Teshnehlab M, Nie K, Su M-Y (2013) Specificity enhancement in classification of breast mri lesion based on multi-classifier. Neural Comput & Applic 22(S1):35–45
Krishnaveni A, Shankar R, Duraisamy S (2021) Versatile duck traveler optimization algorithm using triple segmentation methods for Springer Nature 2021 LATEX template Breast DCE-MRI segmentation using SMAQOBL 43 mammogram image segmentation to improving accuracy. https://doi.org/10.2139/ssrn.3803814
Kumar M, Mehta KK (2011) A texture based tumor detection and automatic segmentation using seeded region growing method. Int J Comp Tech Appl 2:855–859
Levman J, Leung T, Causer P, Plewes D, Martel AL (2008) Classification of dynamic contrast-enhanced magnetic resonance breast lesions by support vector machines. IEEE Trans Med Imaging 27(5):688–696
Li S, Chen H, Wang M, Heidari AA, Mirjalili S (2020) Slime mould algorithm: a new method for stochastic optimization. Futur Gener Comput Syst 111:300–323
Lingle W, Erickson BJ, Zuley ML, Jarosz R, Bonaccio E (2007) Filippini J, Radiology Data from the Cancer Genome Atlas Breast Invasive Carcinoma Collection [TCGA-BRCA], Gruszauskas N
Mann RM, Kuhl CK, Kinkel K, Boetes C (2008) Breast MRI: Guidelines from the european society of breast imaging. Eur Radiol 18:1307–1318
ME GM, Subashini MM (2019) Medical imaging with intelligent systems: a review Sangaiah, A.K. (ed.) Deep Learning and Parallel Computing Environment for Bioengineering Systems, pp 53–73. Academic Press Chap 4. https://doi.org/10.1016/B978-0-12-816718-2.00011-7
Mirjalili S (2016) Dragonfly algorithm: a aew meta-heuristic optimization technique for solving single-objective, discrete, and multi-objective problems. Neural Comput and Applic 27:1053–1073
Mirjalili S, Mirjalili SM, Hatamlou A (2016) Multi-verse optimizer: a nature-inspired algorithm for global optimization. Natural Computing and Applications 27:495–513
Mirjalili SZ, Mirjalili S, Saremi S, Faris H, Aljarah I (2018) Grasshopper optimization algorithm for multi-objective optimization problems. Appl Intell 48:805–820
Mohan J, Krishnavenib V, Guo Y (2014) A survey on the magnetic resonance image denoising methods. Biomed Signal Process Control 9:56–69
Mustra M, Grgic M (2013) Robust automatic breast and pectoral mus- cle segmentation from scanned mammograms. Signal processing 93 (10):2817–2827
Naidu MSR, Kumar PR, Chiranjeevi K (2018) Shannon and Fuzzy entropy based evolutionary image thresholding for image segmentation. Alex Eng J 57:1643–1655
Nie K, Chen J-H, Chan S, Chau M-K I, Yu HJ, Bahri S, Tseng T, Nalcioglu O, Su M-Y (2008) Development of a quantitative method for analysis of breast density based on three-dimensional breast mri. Med Phys 35:5253–5262
Oliver A, Freixenet J, Marti J, Perez E, Pont J, Denton ERE, Zwiggelaar R (2010) A review of automatic mass detection and segmentation in mammographic images. Med Image Anal 14:87–110
Patra DK, Si T, Mondal S, Mukherjee P (2021) Breast DCE-MRI segmentation for lesion detection by multi-level thresholding using student psychological based optimization, biomedical signal processing and control 69(102925). https://doi.org/10.1016/j.bspc
Patra DK, Mondal S, Mukherjee P (2021) Grammatical fireworks algorithm method for breast lesion segmentation in DCE-MRI. International Journal of Innovative Technology and Exploring Engineering 10(7):170–182. https://doi.org/10.35940/ijitee.G9054.0510721
Perona P, Malik J (1990) Scale-space and edge detection using Anisotropic Diffussion. IEEE Trans. Pattern Anal Mach Intell 12(7):629–639
Piantadosi G, Marrone S, Galli A, Sansone M, Sansone C (2019) DCE-MRI Breast Lesions Segmentation with a 3TP U-Net Deep Convolutional Neural Network. In: 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS)
Pizer SM, Johnston RE, Ericksen JP, Yankaskas BC, Muller KE (2002) Contrast-limited Adaptive Histogram Equalization, Speed and Effectiveness. In: 1990 Proceedings of the First Conference on Visualization in Biomedical Computing. https://doi.org/10.1109/VBC.1990.109340
Rahnamayan S (2007) Quasi-Oppositional Differential Evolution. IEEE Congress on Evolutionary Computation. https://doi.org/10.1109/CEC.2007.4424748, Salama, MMA
Samantaray L, Hembram S, Panda R (2020) A new Harris Hawks-Cuckoo search optimizer for multilevel thresholding of thermogram images. International Information and Engineering Technology Association 34:541–551. https://doi.org/10.18280/ria.340503
Shannon C, Weaver W (1964) The mathematical theory of communication. Urbana Ill, University of illinois press
Suradi SH, Abdullah KA, Is MI (2021) Breast lesions detection using FADHECAL and multilevel otsu thresholding segmentation in digital mammograms. In: Badnjevic A, Gurbeta Pokvi CL (eds) Proceedings of the International Conference on Medical and Biological Engineering, CMBEBIH 2021, April 21–24, 2021, Mostar, Bosnia and Herzegovina. CMBEBIH 2021, vol 84. Springer. https://doi.org/10.1007/978-3-030-73909-685
Shi J, Sahiner B, Chan HP, Ge J, Hadjiiski L, Helvie MA, Nees A, Wu YT, Wei J, Zhou C, Zhang Y, Cui J (2008) Characterization of mammographic masses based on level set segmentation with new image features and patient information. Med Phys 35:280–290
Si T, De A, Bhattacharjee AK (2015) Brain mri segmentation for tumor detection via entropy maximization using grammatical swarm. Interna- tional Journal of Wavelets Multiresolution and Information Processing 13(5):1–32
Si T, De A, Bhattacharjee AK (2015) Brain mri segmentation for tumor detection via entropy maximization using grammatical swarm. Interna- tional Journal of Wavelets, Multiresolution and Information Processing 13(5). https://doi.org/10.1142/S0219691315500393
Si T, Dutta R (2019) Partial opposition-based particle swarm optimizer in artificial neural network training for medical data classification. Int J Inf Technol Decis Mak 18(5):1717–1750
Si T, Miranda P, Galdino JV, Nascimento A (2021) Grammar-based automatic programming for medical data classification: an experimental study. Artif Intell Rev, 54(3). https://doi.org/10.1007/s10462-020-09949-9
Thakran S, Chatterjee S, Singhal M, Gupta RK, Singh A (2018) Automatic outer and inner breast tissue segmentation using multi-parametric mri images of breast tumor patients. Plos One 13(1):e0190348. https://doi.org/10.1371/journal.pone.0190348
Tharwat A (2018) Classification assessment methods. Applied Computing and Informatics 17:168–192
(2019) The Cancer Imaging Archive: TCGA-BRCA https://wiki.cancerimagingarchive.net/display/Public/TCGA-BRCA. Accessed 02 Mar 2019
Trelea IC (2002) The particle swarm optimization algorithm: Convergence analysis and parameter selection. Inf Process Lett 85:317–325
Tizhoosh HR (2005) Opposition-Based Learning: A New Scheme for Machine Intelligence, I. pp 695–701
Triantaphyllou E (2000) Multi-Criteria Decision Making Methods: A Comparative Study 44, 3rd edn. Springer. https://doi.org/10.1007/978-1-4757-3157-6
Tukey JW (1949) Comparing individual means in the analysis of variance. Biometrics 5(2):99–114
Tuncay AH, Akduman I (2015) Realistic microwave breast models through t1-weighted 3-d mri data. IEEE Trans Biomed Eng 62(2):688–698
Wajid SK, Hussain A, Huang K (2018) Three-dimensional local energy-based shape histogram (3d-lesh)-based feature extraction– a novel tech- nique. Expert Syst Appl 112:388–440
Wang H, Zhang Q, Wang Y, Hu H (2018) Structured probabilistic pruning for convolutional neural network acceleration. In: Proceedings of the British Machine Vision Conference (BMVC) v3
World Health Organization (WHO) (2021) Breast cancer. https://www.who.int/news-room/fact-sheets/detail/breast-cancer. Accessed 08 Feb 2021
Wu Q, Salganicoff M, Krishnan A, Fussell DS, Markey MK (2006) Inter-active lesion segmentation on dynamic contrast enhanced breast mri using a markov model. Proceedings Volume 6144, Medical Imaging 2006: Image Processing; 61444M
Xu X, Fu L, Chen Y, Larsson R, Zhang D, Suo S, Hua J, Zhao J (2018) Breast region segmentation being convolutional neural network in dynamic contrast enhanced MRI. 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society 750–753
Yao J, Chen J, Chow C (2009) Breast tumor analysis in dynamic contrast enhanced mri using texture features and wavelet transform. IEEE J Sel Top Signal Process 3:94–100
Zhang H, Foo SW, Krishnan SM, Hua Thng C (2004) Computer aided detection of breast masses from digitized mammograms, IEEE International Workshop on Biomedical Circuits and Systems :1–4
Funding
Not Applicable.
Author information
Authors and Affiliations
Contributions
Dipak Kumar Patra Conceptualization of this study, Methodology, Programming, Writing, and Editing. Tapas Si Conceptualization of this study, Methodology, Data collection, Programming, Writing - Original draft preparation, Review, Editing. Sukumar Mondal Conceptualization of this study, Writing, Review, Editing. Prakash Mukherjee Conceptualization of this study, Writing, Review, Editing.
Corresponding author
Ethics declarations
Ethics approval
Not Applicable.
Consent to participate
Not Applicable.
Conflict of Interests/Competing interests
Authors declared that there are no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tapas Si, Sukumar Mondal and Prakash Mukherjee are contributed equally to this work.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Patra, D.K., Si, T., Mondal, S. et al. Breast lesion detection from MRI images using quasi-oppositional slime mould algorithm. Multimed Tools Appl 82, 30599–30641 (2023). https://doi.org/10.1007/s11042-023-14329-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14329-w