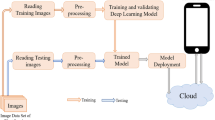
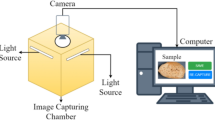
Abstract
In own global economy, the agricultural sector plays a pivotal role in every aspect of our modern life. One of the most important issues that matters in agriculture, that leads to huge economic losses, are the crop diseases. The reliable and accurate diagnosis of plant diseases, even today, remains one of the most difficult tasks. An efficient, accurate and rapid diagnosis of plant disease is active area of research. One of the solutions that has been proposed is Deep Learning (DL). DL is a vital approach in many fields, including agriculture, as it has the potential to reach a high level of accuracy and efficiency. Various authors have investigated DL techniques for agriculture, but most of them examine a very limited dataset or few models and optimizers. In constrast with existing publications, we have performed the most thoroughly examination of all the state of the art DL models resulting in discovering the best models and parameters for utilizing DL in modern agricalture. The experimental results have shown that the DenseNet201 model in combination with the Adam optimization algorithm achieves the highest testing accuracy score of 99.87% surpassing all other DL architectures.










Similar content being viewed by others
References
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M et al (2016) Tensorflow: large-scale machine learning on heterogeneous distributed systems. arXiv:1603.04467
Amodei D, Ananthanarayanan S, Anubhai R, Bai J, Battenberg E, Case C, Casper J, Catanzaro B, Cheng Q, Chen G et al (2016) Deep speech 2: end-to-end speech recognition in english and mandarin. In: International conference on machine learning, pp 173–182
Arsenovic M, Karanovic M, Sladojevic S, Anderla A, Stefanovic D (2019) Solving current limitations of deep learning based approaches for plant disease detection. Symmetry 11(7):939
Atila Ü, Ua̧r M, Akyol K, Ua̧r E (2021) Plant leaf disease classification using efficientnet deep learning model. Ecolog Inform 61:101182
Brahimi M, Arsenovic M, Laraba S, Sladojevic S, Boukhalfa K, Moussaoui A (2018) Deep learning for plant diseases: detection and saliency map visualisation. In: Human and machine learning, pp 93–117. Springer
Brahimi M, Boukhalfa K, Moussaoui A (2017) Deep learning for tomato diseases: classification and symptoms visualization. Appl Artif Intell 31 (4):299–315
Carranza-Rojas J, Goeau H, Bonnet P, Mata-Montero E, Joly A (2017) Going deeper in the automated identification of herbarium specimens. BMC Evol Biol 17(1):1–14
Chao X, Sun G, Zhao H, Li M, He D (2020) Identification of apple tree leaf diseases based on deep learning models. Symmetry 12(7):1065
Chen J, Chen J, Zhang D, Sun Y, Nanehkaran YA (2020) Using deep transfer learning for image-based plant disease identification. Comput Electron Agric 173:105393
Chen J, Wang W, Zhang D, Zeb A, Nanehkaran YA (2021) Attention embedded lightweight network for maize disease recognition. Plant Pathol 70(3):630–642
Chen J, Zhang D, Nanehkaran YA (2020) Identifying plant diseases using deep transfer learning and enhanced lightweight network. Multimed Tools Appl 79(41):31497–31515
Chen J, Zhang D, Nanehkaran YA, Li D (2020) Detection of rice plant diseases based on deep transfer learning. J Sci Food Agric 100(7):3246–3256
Chetlur S, Woolley C, Vandermersch P, Cohen J, Tran J, Catanzaro B, Shelhamer E (2014) cudnn: efficient primitives for deep learning. arXiv:1410.0759
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258
Ciresan D, Giusti A, Gambardella L, Schmidhuber J (2012) Deep neural networks segment neuronal membranes in electron microscopy images. Adv Neural Inform Process Syst 25:2843–2851
Collobert R, Weston J (2008) A unified architecture for natural language processing: deep neural networks with multitask learning. In: Proceedings of the 25th international conference on Machine learning, pp 160–167
Deng L, Yu D (2014) Deep learning: methods and applications. Found Trends Signal Process 7(3–4):197–387
Dozat T (2016) Incorporating nesterov momentum into adam. In: International conference on learning representations
Duchi J, Hazan E, Singer Y (2011) Adaptive subgradient methods for online learning and stochastic optimization. Journal of Machine Learning Research 12(7)
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Comput Electron Agric 145:311–318
Gadekallu TR, Rajput DS, Reddy M, Lakshmanna K, Bhattacharya S, Singh S, Jolfaei A, Alazab M (2021) A novel pca–whale optimization-based deep neural network model for classification of tomato plant diseases using gpu. J Real-Time Image Proc 18(4):1383–1396
Geetharamani G, Pandian A (2019) Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput Electric Eng 76:323–338
Goutte C, Gaussier E (2005) A probabilistic interpretation of precision, recall and f-score, with implication for evaluation. In: European conference on information retrieval, pp 345–359. Springer
Hassan SM, Maji AK, Jasiński M, Leonowicz Z, Jasińska E (2021) Identification of plant-leaf diseases using cnn and transfer-learning approach. Electronics 10(12):1388
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
He K, Zhang X, Ren S, Sun J (2016) Identity mappings in deep residual networks. In: European conference on computer vision, pp 630–645. Springer
Hinton G, Srivastava N, Swersky K (2012) Neural networks for machine learning lecture 6a overview of mini-batch gradient descent. Cited on 14(8)
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Hughes D, Salathé M, et al. (2015) An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv:1511.08060
Kamal K, Yin Z, Wu M, Wu Z (2019) Depthwise separable convolution architectures for plant disease classification. Comput Electron Agric 165:104948
Khirade SD, Patil A (2015) Plant disease detection using image processing. In: 2015 International conference on computing communication control and automation, pp 768–771. IEEE
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv:1412.6980
Krizhevsky A, Sutskever I, Hinton G (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Lee SH, Goëau H, Bonnet P, Joly A (2020) New perspectives on plant disease characterization based on deep learning. Comput Electron Agric 170:105220
Liakos KG, Busato P, Moshou D, Pearson S, Bochtis D (2018) Machine learning in agriculture: a review. Sensors 18(8):2674
Lin M, Chen Q, Yan S (2013) Network in network. arXiv:1312.4400
Maeda-Gutiérrez V, Galván-Tejada CE, Zanella-Calzada LA, Celaya-Padilla JM, Galván-Tejada JI, Gamboa-Rosales H, Luna-García H, Magallanes-Quintanar R, Guerrero Méndez CA, Olvera-Olvera CA (2020) Comparison of convolutional neural network architectures for classification of tomato plant diseases. Appl Sci 10(4):1245
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1419
Rawat W, Wang Z (2017) Deep convolutional neural networks for image classification: a comprehensive review. Neur Comput 29(9):2352–2449
Ruder S (2016) An overview of gradient descent optimization algorithms. arXiv:1609.04747
Saleem MH, Potgieter J, Arif KM (2020) Plant disease classification: a comparative evaluation of convolutional neural networks and deep learning optimizers. Plants 9(10):1319
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: iresiduals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, Stefanovic D (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Computational Intelligence and Neuroscience, 2016
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):1929–1958
Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2016) Inception-v4, inception-resnet and the impact of residual connections on learning. arXiv:1602.07261
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Too EC, Yujian L, Njuki S, Yingchun L (2019) A comparative study of fine-tuning deep learning models for plant disease identification. Comput Electron Agric 161:272–279
Wu H, Prasad S (2017) Semi-supervised deep learning using pseudo labels for hyperspectral image classification. IEEE Trans Image Process 27 (3):1259–1270
Zeiler MD (2012) Adadelta: an adaptive learning rate method. arXiv:1212.5701
Zhang P, Yang L, Li D (2020) Efficientnet-b4-ranger: a novel method for greenhouse cucumber disease recognition under natural complex environment. Comput Electron Agric 176:105652
Zhang X, Qiao Y, Meng F, Fan C, Zhang M (2018) Identification of maize leaf diseases using improved deep convolutional neural networks. IEEE Access 6:30370–30377
Zoph B, Vasudevan V, Shlens J, Le QV (2018) Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 8697–8710
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sanida, T., Tsiktsiris, D., Sideris, A. et al. A heterogeneous implementation for plant disease identification using deep learning. Multimed Tools Appl 81, 15041–15059 (2022). https://doi.org/10.1007/s11042-022-12461-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-12461-7