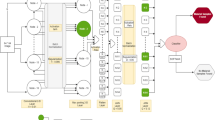
Abstract
Automated malaria parasite detection in thin-blood smear images is an important way to improve the diagnostic performance. Although deep convolutional neural networks (DCNNs) perform well in many image classification tasks, accurate classification of malaria parasite remains challenging due to the lack of training data, poor quality of smears and inter-class similarity. In this paper, we present a novel hybrid model dubbed RAL-CNN-SVM, which is composed of multiple residual attention learning convolution neural network (RAL-CNN) modules, a global average pooling (GAP) block and a classifier trained by a support vector machine (SVM). Each RAL-CNN block consists of residual learning and a new attention mechanism, which is mainly used to extract image depth activation features. The classification layer takes advantage of strong points of the SVM classification algorithm, solves the problem of nonlinear separable and improves the detection accuracy for malaria parasites. During our experiments, we evaluate the proposed RAL-CNN-SVM model on public Malaria Cell Images dataset, and the results show that the accuracy of the proposed novel hybrid model is 99.7%. We also visualized the class activation mapping (CAM) obtained by RAL-CNN50-SVM and ResNet50. The experimental results show that the RAL-CNN50-SVM (a single 50-layer model) model has strong attention ability and can highlight parasitic cells rather than background tissues in thin-blood smear images.











Similar content being viewed by others
References
Ba J L, Kiros J R, Hinton G E (2016) Layer Normalization [J]. Available: https://arxiv.org/abs/1607.06450
Bibin D, Nair MS, Punitha P (2017) Malaria parasite detection from peripheral blood smear images using deep belief networks. IEEE Access 5:9099–9108
Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2018) DeepLab: semantic image segmentation with deep convolutional nets, Atrous convolution, and fully connected CRFs. IEEE Transactions on Pattern Analysis & Machine Intelligence 40(4):834–848
Corbetta M, Shulman G (2002) Control of goal-directed and stimulus-driven attention in the brain. Nat Rev Neurosci 3(1):201–215
Das DK, Ghosh M, Pal M, Maiti AK, Chakraborty C (2013) Machine learning approach for automated screening of malaria parasite using light microscopic images. Micron 45:97–106
Diaz M, Ferrer MA, Impedovo D, Pirlo G, Vessio G (2019) Dynamically enhanced static handwriting representation for Parkinson's disease detection [J]. Pattern Recogn Lett 128:204–210
Dong Y et al (2017) Evaluations of deep convolutional neural networks for automatic identification of malaria infected cells. In: 2017 IEEE EMBS international conference on Biomedical & Health Informatics (BHI), Orlando, FL, 2017, pp 101–104
Dou H, Deng Y, Yan T, Wu H, Lin X, Dai Q (2020) Residual D2NN: training diffractive deep neural networks via learnable light shortcuts [J]. Opt Lett 45(10):2688–2691
Gopakumar GP, Swetha M et al (2017) Convolutional neural network-based malaria diagnosis from focus stack of blood smear images acquired using custom-built slide scanner. J Biophotonics 10(3):162–171
Gutman JR, Lucchi NW, Cantey PT, Steinhardt LC, Samuels AM, Kamb ML, Kapella BK, McElroy PD, Udhayakumar V, Lindblade KA (2020) Malaria and parasitic neglected tropical diseases: potential Syndemics with COVID-19?[J]. Am J Trop Med Hyg 103(2)
Han J, Yanrong G (2020) Multi-class multimodal semantic segmentation with an improved 3D fully convolutional networks. Neurocomputing 91:220–226
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proc. IEEE Conf. Comput. Vis. Pattern Recognit, pp 770–778
Hinton GE (2002) Training products of experts by minimizing contrastive divergence. Neural Comput 14(8):1771–1800
Hinton GE (2010) Rectified linear units improve restricted Boltzmann machines Vinod Nair. In: International conference on international conference on machine Learning. Omnipress
Hossain MS, Al-Hammadi M, Muhammad G (2019) Automatic fruit classification using deep learning for industrial applications. IEEE Trans Ind Inform 15(2):1027–1034
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks in proc. In: IEEE Conf. Comput. Vis. Pattern Recogn, vol 2018, pp 7132–7141
Huang Y, Xu H (2021) Fully convolutional network with attention modules for semantic segmentation [J]. Signal Image Video Process 15:1–9
Huang G, Liu Z, Maaten LVD et al (2017) Densely connected convolutional networks. In: IEEE Conference on Computer Vision & Pattern Recognition
Ioffe S, Szegedy C (2015) Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift [J]. 45(2):204–210
Jie H, Li S, Albanie S et al (2017) Squeeze-and-excitation networks. IEEE Trans Pattern Anal Mach Intell 99(1):1–1
Kowalski M, Naruniec J, Trzcinski T (2017) Deep alignment network: a convolutional neural network for robust face alignment. In: IEEE Conference on Computer Vision & Pattern Recognition Workshops
Krizhevsky, Alex, Sutskever, et al (2017) ImageNet classification with deep convolutional neural networks [J]. Commun ACM, 2017: 1106–1114.
Larochelle H, Hinton GE (2010) Learning to combine foveal glimpses with a third-order Boltzmann machine. In: Advances in Neural Information Processing Systems 23: 24th Annual Conference on Neural Information Processing Systems
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86:2278–2323
Li G, Li L, Zhu H, Liu X, Jiao L (2019) Adaptive multiscale deep fusion residual network for remote sensing image classification. IEEE Trans Geoence Remote Sens 20(1):1–16
Liang Z, Powell A, Ersoy I, Poostchi M, Silamut K et al (2016) CNN-based image analysis for malaria diagnosis. In: Proceedings—2016 IEEE international conference on bioinformatics and biomedicine, BIBM 2016. IEEE, Piscataway, pp 493–496
Lin M, Chen Q, Yan S (2013) Network in network. Comput Sci
Lin TY, Goyal P, Girshick R et al (2017) Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell 99(1):2999–3007
Liu S, Luo H, Tu Y et al (2018) Wide Contextual Residual Network with Active Learning for Remote Sensing Image Classification. In: IGARSS 2018–2018 IEEE International Geoscience and Remote Sensing Symposium, vol 2018, pp 7145–7148
Luo R, Zhong X, Chen E (2020) Image classification with a MSF dropout [J]. Multimed Tools Appl 79(4):1–11
M.A (2018) World Health Organization (WHO) (2018) World malaria report. Available at http://www.who.int/malaria/publications/world-malaria-report-2017/report/en/. Accessed 8 Dec 2018
Masud M, Alhumyani Sultan S, Cheikhrouhou O et al (2020) Leveraging deep learning techniques for malaria parasite detection using Mobile application. Wirel Commun Mob Comput 2020(2):1530–8669
Rajaraman S, Jaeger S, Antani S (2019) Performance evaluation of deep neural ensembles toward malaria parasite detection in thin-blood smear images. PeerJ
Rajaraman S, Jaeger S, Antani SK (2019) Performance evaluation of deep neural ensembles toward malaria parasite detection in thin-blood smear images [J]. PeerJ 7(1):115–123
Ross NE, Pritchard CJ, Rubin DM, Dusé AG (2006) Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Med Biol Eng Comput 44:427–436
Rui Z, Ruqiang Y, Zhenghua C et al (2019) Deep learning and its applications to machine health monitoring. Mech Syst Signal Process 115(1):213–237
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L (2015) ImageNet large scale visual recognition challenge. Int J Comput Vis 115(3):211–252
Sain SR (1996) The nature of statistical learning theory. Technometrics 38(4):409–409
Selvaraju RR, Cogswell M, Das A et al (2020) Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis 12(2):336–359
Sivaramakrishnan R, Antani SK et al (2018) Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images. PeerJ 6(1):215–223
Srivastava RK, Greff K, Schmidhuber J (2015) Training very deep networks [J]. Computer Science
Suzuki K (2017) Overview of deep learning in medical imaging. Radiol Phys Technol 10:257–273
Swami A, Jain R (2012) Scikit-learn: machine learning in Python. J Mach Learn Res 12(10):2825–2830
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2014) Going deeper with convoltions. ArXiv preprint. arXiv:1409.4842
Wang X, Shrivastava A, Gupta A (2017) A-fast-RCNN: hard positive generation via adversary for object detection. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR), Honolulu, HI, vol 2017, pp 3039–3048
Wang X, Shrivastava A, Gupta A (2017) A-fast-RCNN: hard positive generation via adversary for object detection
Wang F, Jiang M , Qian C et al (2017) Residual attention network for image classification
Weese J, Lorenz C (2016) Four challenges in medical image analysis from an industrial perspective. Med. Image Anal 33:44–49
Woo S, Park J, Lee JY et al (2018) CBAM: convolutional block attention module. Springer, Cham
Wu H, Li D, Cheng M (2019) Source: International Journal of Intelligent Information and Database Systems. 12(3):212–228
Xie Y, Xia Y, Zhang J, Song Y, Feng D, Fulham M, Cai W (2019) Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans Med Imaging 38(4):991–1004
Xing L, Schreibmann E, Levy D et al (2017) Multiscale Image Registration. Mathematical Biosciences and Engineering (Online) 3(2):389–418
Xiong W, Du B, Zhang L et al (2016) Regularizing deep convolutional neural networks with a structured decorrelation constraint. In: 2016 IEEE 16th international conference on data mining (ICDM)
Yang, Yifan & Huang, Qijing & Wu, Bichen & Zhang et al (2018) Synetgy: Algorithm-hardware Co-design for ConvNet Accelerators on Embedded FPGAs
Yu L, Chen H, Dou Q, Qin J, Heng PA (2017) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imag 36(4):94–1004
Zhang Q, Bai C, Liu Z, Yang LT, Yu H, Zhao J, Yuan H (2020) A GPU-based residual network for medical image classification in smart medicine. Inf Sci 536(1):91–100
Zhong X, Gong O, Huang W et al (2020) Multi-scale residual network for image classification. In: IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE
Acknowledgements
We acknowledge the official NIH Website for the Public Malaria Cell Images dataset to support our research work. Meanwhile, we also acknowledge the National Natural Science Foundation of China (No.61370075) for funding our research work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, D., Ma, Z. Residual attention learning network and SVM for malaria parasite detection. Multimed Tools Appl 81, 10935–10960 (2022). https://doi.org/10.1007/s11042-022-12373-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-12373-6