Abstract
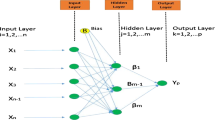
White blood cells (Leukocytes) are considered to be an essential part of the human body’s immune system. The count of WBCs is considered to be a parameter for the indication of disease. Over time several methods have been proposed to classify these WBCs into their subtypes namely Neutrophils, Eosinophils, Basophils, Lymphocytes, and Monocytes which helps in the estimation of the body’s WBC count. These methods range from various morphological image processing-based methodologies to advanced deep neural systems. Due to the superior ability of neural systems to achieve the state of the art results more research is been carried out in this field. However, most of the such previously proposed methods have concentrated only in establishing and explaining the overall methodology for achieving high accuracy scores and less emphasis has been made in discussing the impact of modular changes in such methodologies like the impact of various activation functions, optimizers and data pre-processing methods very explicitly for this problem. This has led to a deficiency of work to be carried out with very recently developed activation functions and more essentially optimization algorithms other than backpropagation. It is extremely essential to explore and analyse different modules of the methodology to accelerate future research work further which might possibly help the research community in achieving a much better and efficient solution. This paper compares various architectures and discusses the behaviour and impact of different hyperparameters and proposes a novel methodology by incorporating recently developed swish activation to enhance the results. Unlike previously proposed methods of proposing single better neural network model this paper suggests a good choice of modular changes that could be incorporated in future works to enhance their results.
















Similar content being viewed by others
References
Xing F, Yang L (2016) Robust nucleus/cell detection and segmentation in digital pathology and microscopy images: a comprehensive review. IEEE Rev Biomed Eng 9:234–263. https://doi.org/10.1109/rbme.2016.2515127
Moshavash Z, Danyali H, Helfroush MS (2018) An automatic and robust decision support system for accurate acute leukemia diagnosis from blood microscopic images. J Digit Imaging 31(5):702–717. https://doi.org/10.1007/s10278-018-0074-y
Irshad H, Veillard A, Roux L, Racoceanu D (2014) Methods for nuclei detection, segmentation, and classification in digital histopathology: a review—current status and future potential. IEEE Rev Biomed Eng 7:97–114. https://doi.org/10.1109/rbme.2013.2295804
Razzak, M. I., & Naz, S. (2017). Microscopic blood smear segmentation and classification using deep contour aware CNN and extreme machine learning. 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). doi: https://doi.org/10.1109/cvprw.2017.111
Miotto R, Wang F, Wang S, Jiang X, Dudley JT (2017) Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform 19(6):1236–1246. https://doi.org/10.1093/bib/bbx044
Shahin A, Guo Y, Amin K, Sharawi AA (2019) White blood cells identification system based on convolutional deep neural learning networks. Comput Methods Prog Biomed 168:69–80. https://doi.org/10.1016/j.cmpb.2017.11.015
Kutlu H, Avci E, Özyurt F (2020) White blood cells detection and classification based on regional convolutional neural networks. Med Hypotheses 135:109472. https://doi.org/10.1016/j.mehy.2019.109472
Sharma M, Bhave A, Janghel RR (2019) White blood cell classification using convolutional neural network. Advances in Intelligent Systems and Computing Soft Computing and Signal Processing:135–143. https://doi.org/10.1007/978-981-13-3600-3_13
Zhao J, Zhang M, Zhou Z, Chu J, Cao F (2016) Automatic detection and classification of leukocytes using convolutional neural networks. Medical & Biological Engineering & Computing 55(8):1287–1301. https://doi.org/10.1007/s11517-016-1590-x
Hegde RB, Prasad K, Hebbar H, Singh BMK (2019) Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybernetics and Biomedical Engineering 39(2):382–392. https://doi.org/10.1016/j.bbe.2019.01.005
Qi X, Xiao R, Li C-G, Qiao Y, Guo J, Tang X (2014) Pairwise rotation invariant co-occurrence local binary pattern. IEEE Trans Pattern Anal Mach Intell 36(11):2199–2213. https://doi.org/10.1109/tpami.2014.2316826
Breiman L (2001) Mach Learn 45(1):5–32. https://doi.org/10.1023/a:1010933404324
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60(6):84–90. https://doi.org/10.1145/3065386
Lecun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324. https://doi.org/10.1109/5.726791
Redmon, Joseph, Farhadi, Ali. (2018, April 8). YOLOv3: An Incremental Improvement. Retrieved from https://arxiv.org/abs/1804.02767
Liu, Wei, Anguelov, Dragomir, Erhan, Dumitru, … Reed. (2016, December 29). SSD: Single Shot MultiBox Detector. Retrieved from https://arxiv.org/abs/1512.02325v5
Girshick, Ross, Donahue, Jeff, Darrell, Trevor, … Jitendra. (2014, October 22). Rich feature hierarchies for accurate object detection and semantic segmentation. Retrieved from https://arxiv.org/abs/1311.2524
Girshick, Ross. (2015, September 27). Fast R-CNN. Retrieved from https://arxiv.org/abs/1504.08083
Ren S, He K, Girshick R, Sun J (2017) Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell 39(6):1137–1149. https://doi.org/10.1109/tpami.2016.2577031
Yu, W., Chang, J., Yang, C., Zhang, L., Shen, H., Xia, Y., & Sha, J. (2017). Automatic classification of leukocytes using deep neural network. 2017 IEEE 12th International Conference on ASIC (ASICON). doi: https://doi.org/10.1109/asicon.2017.8252657
Zhang, Ren, Sun, Jian. (2015, December 10). Deep residual learning for image recognition. Retrieved from https://arxiv.org/abs/1512.03385
Simonyan, Karen, Zisserman, Andrew. (2015, April 10). Very deep convolutional networks for large-scale image recognition. Retrieved from https://arxiv.org/abs/1409.1556
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., & Wojna, Z. (2016). Rethinking the inception architecture for computer vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). doi: https://doi.org/10.1109/cvpr.2016.308
Chollet, François. (2017, April 4). Xception: Deep Learning with Depthwise Separable Convolutions. Retrieved from https://arxiv.org/abs/1610.02357
Qin F, Gao N, Peng Y, Wu Z, Shen S, Grudtsin A (2018) Fine-grained leukocyte classification with deep residual learning for microscopic images. Comput Methods Prog Biomed 162:243–252. https://doi.org/10.1016/j.cmpb.2018.05.024
Motlagh, M. H., Jannesari, M., Rezaei, Z., Totonchi, M., & Baharvand, H. (2018). Automatic white blood cell classification using pre-trained deep learning models: ResNet and inception. Tenth International Conference on Machine Vision (ICMV 2017). doi: https://doi.org/10.1117/12.2311282
Jung, Mohammed, Alikhanov, Jumabek, Mohaisen, Aziz, … Dae Hun. (2019, October 2). W-Net: A CNN-based Architecture for White Blood Cells Image Classification. Retrieved from https://arxiv.org/abs/1910.01091
Rezatofighi SH, Soltanian-Zadeh H (2011) Automatic recognition of five types of white blood cells in peripheral blood. Comput Med Imaging Graph 35(4):333–343. https://doi.org/10.1016/j.compmedimag.2011.01.003
Ramachandran, Zoph, Barret, V. Q. (2017, October 27). Searching for activation functions. Retrieved from https://arxiv.org/abs/1710.05941
Ramachandran, Zoph. (n.d.). Swish: a Self-Gated Activation Function - arxiv.org. Retrieved from https://arxiv.org/pdf/1710.05941v1.pdf
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359. https://doi.org/10.1109/tkde.2009.191
Kingma, P. D., Jimmy, Ba. (2017, January 30). Adam: A Method for Stochastic Optimization. Retrieved from https://arxiv.org/abs/1412.6980
Jain, Kar, & Purushottam. (2017, December 21). Non-convex optimization for machine learning. Retrieved from https://arxiv.org/abs/1712.07897
Sergey. (2015, March 2). Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Retrieved from https://arxiv.org/abs/1502.03167
Shuai Liu, Xinyu Liu, Shuai Wang, et al. Fuzzy-Aided Solution for Out-of-View Challenge in Visual Tracking under IoT Assisted Complex Environment. Neural Computing and Applications, 2020, doi: https://doi.org/10.1007/s00521-020-05021-3
Shuai Liu, Chunli Guo, Fadi Al-Turjman, et al. Reliability of response region: A novel mechanism in visual tracking by edge computing for IIoT environments, Mechanical Systems and Signal Processing, 2020, 138, 106537
Liu S, Liu G, Zhou H (2019) A robust parallel object tracking method for illumination variations. Mobile Networks and Applications 24(1):5–17
Author information
Authors and Affiliations
Contributions
B A Harshanand: Designed and performed experiments, analysed data and wrote the paper.
Arun Kumar Sangaiah: Reviewed the paper and helped in drafting it.
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Harshanand, B.A., Sangaiah, A.K. Comprehensive Analysis of Deep Learning Methodology in Classification of Leukocytes and Enhancement Using Swish Activation Units. Mobile Netw Appl 25, 2302–2320 (2020). https://doi.org/10.1007/s11036-020-01614-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11036-020-01614-3