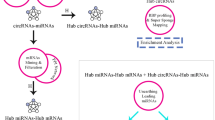
Abstract
Background
Circular RNAs (circRNAs) take an effect on tumorigenesis and progression. However, circRNAs have not been systematically identified in breast cancer (BC) as crucial regulators in multitudinous biological processes. This study is conducted to explore novel circRNAs in BC and the corresponding mechanisms of their action.
Methods
The circRNA expression profile and RNA-sequencing data about BC were respectively downloaded from public database. Differentially expressed circRNAs, miRNAs, and mRNAs were identified by fold change filtering. The competing endogenous RNAs (ceRNAs) network was established based on the relationship between circular RNAs, miRNAs and mRNAs. GO and KEGG enrichment analysis of the overlapped genes were carried out to predict the potential functions and mechanisms of circRNAs in BC. The CytoHubba plugin in Cytoscape was applied to identify the hub genes from the PPI regulatory network. Kaplan–Meier plotter was used to perform survival analysis of these hub genes further. Real-time PCR was performed to test the expression of circRNA in BC tissues. Cell function studies including transwell analysis and CCK-8 analysis were used to investigate circRNAs’ biological functions.
Results
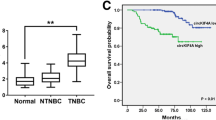
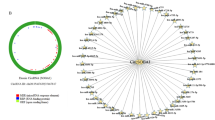
A total of seven circRNAs exhibiting differential expression were identified in this study. After the intersection between the predicted target miRNA and the down-regulated differential miRNAs (DEmiRNAs), circRNA-miRNA interactions involving 3 circRNAs and 4 miRNAs were identified. Venn diagram was utilized to intersect the predicted target genes of the 4 miRNAs and the down-regulated differential genes in BC, and 149 overlapped genes were screened out ulteriorly. Additionally, we built a protein-protein interaction (PPI) network and selected six hub genes. Moreover, the survival data of BC patients suggested that low expression of ADIPOQ, LPL and LEP were significantly correlated with poor prognosis. Results from real-time PCR indicated that hsa_circ_0000375 was significantly down-regulated in breast cancer tissues. Functional in vitro experiments showed that over-expression of hsa_circ_0000375 can restrain proliferation, migration and invasion abilities of breast cancer cells. Further verification indicated that hsa_circ_0000375 exerted its anti-oncogene effect via sponge of miR-7706.
Conclusions
This study constructed and analyzed a circRNA-associated ceRNA regulatory network and uncovered that hsa_circ_0000375 exerted its anti-oncogene effect via sponge of miR-7706.







Similar content being viewed by others
Data Availability
All data in this study are available from the corresponding author upon request.
References
Cardoso F, Paluch-Shimon S, Senkus E, Curigliano G, Aapro MS et al (2020) 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Ann Oncol 31:1623–1649
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388
Chen Q, Wang H, Li Z, Li F, Liang L et al (2022) Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J Hepatol 76:135–147
Wu P, Hou X, Peng M, Deng X, Yan Q et al (2023) Circular RNA circRILPL1 promotes nasopharyngeal carcinoma malignant progression by activating the Hippo-YAP signaling pathway. Cell Death Differ 30:1679–1694
He P, Liu F, Wang Z, Gong H, Zhang M et al (2022) CircKIF4A enhances osteosarcoma proliferation and metastasis by sponging MiR-515-5p and upregulating SLC7A11. Mol Biol Rep 49:4525–4535
Liu L, Gu M, Ma J, Wang Y, Li M et al (2022) CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol Cancer 21:149
Chen J, Yang J, Fei X, Wang X, Wang K (2021) CircRNA ciRS-7: a Novel Oncogene in multiple cancers. Int J Biol Sci 17:379–389
Hu Z, Chen J, Wang M, Weng W, Chen Y et al (2022) Rheumatoid arthritis fibroblast-like synoviocytes maintain tumor-like biological characteristics through ciRS-7-dependent regulation of miR-7. Mol Biol Rep 49:8473–8483
Yu Q, Chen W, Li Y, He J, Wang Y et al (2022) The novel circular RNA HIPK3 accelerates the proliferation and invasion of hepatocellular carcinoma cells by sponging the micro RNA-124 or micro RNA-506/pyruvate dehydrogenase kinase 2 axis. Bioengineered 13:4717–4729
Wang S, Tong H, Su T, Zhou D, Shi W et al (2021) CircTP63 promotes cell proliferation and invasion by regulating EZH2 via sponging miR-217 in gallbladder cancer. Cancer Cell Int 21:608
Huang W, Yang Y, Wu J, Niu Y, Yao Y et al (2020) Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging mir-93-5p to inhibit TGF-beta signalling. Cell Death Differ 27:1709–1727
Li J, Gao X, Zhang Z, Lai Y, Lin X et al (2021) CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes. Mol Cancer 20:138
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L et al (2019) The Landscape of circular RNA in Cancer. Cell 176:869–881e813
Liu F, Sang Y, Zheng Y, Gu L, Meng L et al (2022) circRNF10 regulates tumorigenic Properties and Natural Killer cell-mediated cytotoxicity against breast Cancer through the miR-934/PTEN/PI3k-Akt Axis. Cancers (Basel) 14
Ding F, Lu L, Wu C, Pan X, Liu B et al (2022) circHIPK3 prevents cardiac senescence by acting as a scaffold to recruit ubiquitin ligase to degrade HuR. Theranostics 12:7550–7566
Li Z, Huang C, Bao C, Chen L, Lin M et al (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264
Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D et al (2019) CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol 58:90–99
Pan Z, Zheng J, Zhang J, Lin J, Lai J et al (2022) A Novel protein encoded by Exosomal CircATG4B induces Oxaliplatin Resistance in Colorectal Cancer by promoting Autophagy. Adv Sci (Weinh) 9:e2204513
Song R, Guo P, Ren X, Zhou L, Li P et al (2023) A novel polypeptide CAPG-171aa encoded by circCAPG plays a critical role in triple-negative breast cancer. Mol Cancer 22:104
Liang Y, Cen J, Huang Y, Fang Y, Wang Y et al (2022) CircNTNG1 inhibits renal cell carcinoma progression via HOXA5-mediated epigenetic silencing of slug. Mol Cancer 21:224
Zhong Y, Du Y, Yang X, Mo Y, Fan C et al (2018) Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 17:79
Cortes-Lopez M, Miura P (2016) Emerging functions of circular RNAs. Yale J Biol Med 89:527–537
Ghafouri-Fard S, Hussen BM, Taheri M, Ayatollahi SA (2021) Emerging role of circular RNAs in breast cancer. Pathol Res Pract 223:153496
Yin TF, Zhao DY, Zhou YC, Wang QQ, Yao SK (2021) Identification of the circRNA-miRNA-mRNA regulatory network and its prognostic effect in colorectal cancer. World J Clin Cases 9:4520–4541
Yin TF, Du SY, Zhao DY, Sun XZ, Zhou YC et al (2022) Identification of circ_0000375 and circ_0011536 as novel diagnostic biomarkers of colorectal cancer. World J Clin Cases 10:3352–3368
Su Y, Sun Y, Tang Y, Li H, Wang X et al (2021) Circulating miR-19b-3p as a Novel Prognostic Biomarker for Acute Heart failure. J Am Heart Assoc 10:e022304
Shen Y, Wang L, Wu Y, Ou Y, Lu H et al (2021) A novel diagnostic signature based on three circulating exosomal mircoRNAs for chronic obstructive pulmonary disease. Exp Ther Med 22:717
Wang F, Dai M, Chen H, Li Y, Zhang J et al (2018) Prognostic value of hsa-mir-299 and hsa-mir-7706 in hepatocellular carcinoma. Oncol Lett 16:815–820
Manupati K, Yeeravalli R, Kaushik K, Singh D, Mehra B et al (2021) Activation of CD44-Lipoprotein lipase axis in breast cancer stem cells promotes tumorigenesis. Biochim Biophys Acta Mol Basis Dis 1867:166228
Almanza A, Mnich K, Blomme A, Robinson CM, Rodriguez-Blanco G et al (2022) Regulated IRE1alpha-dependent decay (RIDD)-mediated reprograming of lipid metabolism in cancer. Nat Commun 13:2493
Chung SJ, Nagaraju GP, Nagalingam A, Muniraj N, Kuppusamy P et al (2017) ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis. Autophagy 13:1386–1403
Acknowledgements
No.
Funding
This work was supported by the 2023 Medical science research key project [number 20230148] and the Hebei Provincial Department of Education’s postgraduate innovation ability training funding project. [number CXZZBS2023102].
Author information
Authors and Affiliations
Contributions
H.W. and F.L. conducted data analysis and validation. S.G. and C.G. conceived the study and drafted the manuscript. J.X., Y.L., W.G. and S.Y. performed the experiments. All authors have read and agreed to the final manuscript. S.G. and C.G. confirm the authenticity of all the raw data. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Ethical approval for this study was obtained from the Research Ethics Committee of the Fourth Hospital of Hebei Medical University (number 2019MEC057).
Consent of publication
All the authors have consent to publish.
Consent to participant
All participants included in the study were provided with informed consent.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, H., Liu, F., Xue, J. et al. The investigation of circRNA profiling reveals the regulatory role of the hsa_circ_0000375/miR-7706 pathway in breast cancer. Mol Biol Rep 50, 9993–10004 (2023). https://doi.org/10.1007/s11033-023-08798-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08798-3