Abstract
Background
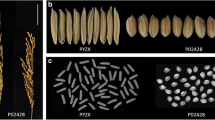
Rice grain chalkiness is an undesirable characteristic that affects grain quality. The aim of this study was to map QTLs controlling grain chalkiness in japonica rice.
Methods and results
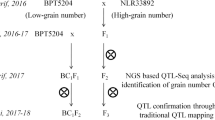
In this study, two japonica rice cultivars with similar grain shapes but different grain chalkiness rates were crossed and the F2 and BC1F2 populations were subjected to QTL-seq analysis to map the QTLs controlling the grain chalkiness rate. QTL-seq analysis revealed SNP index differences on chromosome 1 in both of the segregating populations. Using polymorphic markers between the two parents, QTL mapping was conducted on 213 individual plants in the BC1F2 population. QTL mapping confined a QTL controlling grain chalkiness, qChalk1, to a 1.1 Mb genomic region on chromosome 1. qChalk1 explained 19.7% of the phenotypic variation.
Conclusion
A QTL controlling grain chalkiness qChalk1 was detected in both F2 and BC1F2 segregating populations by QTL-Seq and QTL mapping methods. This result would be helpful for further cloning of the genes controlling grain chalkiness in japonica rice.



Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Liu X, Guo T, Wan X, Wang H, Zhu M, Li A, Su N, Shen Y, Mao B, Zhai H, Mao L, Wan J (2010) Transcriptome analysis of grain-filling caryopses reveals involvement of multiple regulatory pathways in chalky grain formation in rice. BMC Genomics 11:730. https://doi.org/10.1186/1471-2164-11-730
Lin Z, Wang Z, Zhang X, Liu Z, Li G, Wang S, Ding Y (2017) Complementary proteome and transcriptome profiling in developing grains of a notched-belly rice mutant reveals key pathways involved in chalkiness formation. Plant Cell Physiol 58(3):560–573. https://doi.org/10.1093/pcp/pcx001
Tabassum R, Dosaka T, Ichida H, Morita R, Ding Y, Abe T, Katsube-Tanaka T (2020) FLOURY ENDOSPERM11–2 encodes plastid HSP70–2 involved with the temperature-dependent chalkiness of rice (Oryza sativa L.) grains. Plant J 103(2):604–616. https://doi.org/10.1111/tpj.14752
Siebenmorgen TJ, Grigg BC, Lanning SB (2013) Impacts of preharvest factors during kernel development on rice quality and functionality. Annu Rev Food Sci Technol 4(4):101–115. https://doi.org/10.1146/annurev-food-030212-182644
Fitzgerald MA, McCouch SR, Hall RD (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci 14(3):133–139. https://doi.org/10.1016/j.tplants.2008.12.004
Yamakawa H, Hirose T, Kuroda M, Yamaguchi T (2007) Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol 144(1):258–277. https://doi.org/10.1104/pp.107.098665
Tian Z, Qian Q, Liu Q, Yan M, Liu X, Yan C, Liu G, Gao Z, Tang S, Zeng D, Wang Y, Yu J, Gu M, Li J (2009) Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci USA 106(51):21760–21765. https://doi.org/10.1073/pnas.0912396106
Sreenivasulu N, Butardo VM Jr, Misra G, Cuevas RP, Anacleto R, Kavi Kishor PB (2015) Designing climate-resilient rice with ideal grain quality suited for high-temperature stress. J Exp Bot 66(7):1737–1748. https://doi.org/10.1093/jxb/eru544
Dwiningsih Y, Kumar A, Thomas J, Ruiz C, Alkahtani J, Al-Hashimi A, Pereira A (2021) Identification of genomic regions controlling chalkiness and grain characteristics in a recombinant inbred line rice population based on high-throughput SNP markers. Genes 12(11):1690. https://doi.org/10.3390/genes12111690
Ayaad M, Han Z, Zheng K, Hu G, Abo-Yousef M, Sobeih SES, Xing Y (2021) Bin-based genome-wide association studies reveal superior alleles for improvement of appearance quality using a 4-way MAGIC population in rice. J Adv Res 28:183–194. https://doi.org/10.1016/j.jare.2020.08.001
Wang X, Pang Y, Wang C, Chen K, Zhu Y, Shen C, Ali J, Xu J, Li Z (2016) New candidate genes affecting rice grain appearance and milling quality detected by genome-wide and gene-based association analyses. Front Plant Sci 7:1998. https://doi.org/10.3389/fpls.2016.01998
Yang WF, Hao QW, Liang JY, Tan QY, Luan X, Lin SJ, Zhu HT, Bu SH, Liu ZP, Liu GF, Wang SK, Zhang GQ (2022) Fine mapping of two major quantitative trait loci for rice chalkiness with high temperature-enhanced additive effects. Front Plant Sci 13:957863. https://doi.org/10.3389/fpls.2022.957863
Yang W, Xiong L, Liang J, Hao Q, Luan X, Tan Q, Lin S, Zhu H, Liu G, Liu Z, Bu S, Wang S, Zhang G (2021) Substitution mapping of two closely linked QTLs on chromosome 8 controlling grain chalkiness in rice. Rice 14(1):85. https://doi.org/10.1186/s12284-021-00526-4
Qiu XJ, Chen K, Lv WK, Ou XX, Zhu YJ, Xing DY, Yang LW, Fan FJ, Yang J, Xu JL, Zheng TQ, Li ZK (2017) Examining two sets of introgression lines reveals background-independent and stably expressed QTL that improve grain appearance quality in rice (Oryza sativa L). Theor Appl Genet 130(5):951–967. https://doi.org/10.1007/s00122-017-2862-z
Zhu AK, Zhang YX, Zhang ZH, Wang BF, Xue P, Cao YR, Chen YY, Li ZH, Liu QN, Cheng SH, Cao LY (2018) Genetic dissection of qPCG1 for a quantitative trait locus for percentage of chalky grain in rice (Oryza sativa L). Front Plant Sci 9:1173. https://doi.org/10.3389/fpls.2018.01173
Gao Y, Liu C, Li Y, Zhang A, Dong G, Xie L, Zhang B, Ruan B, Hong K, Xue D, Zeng D, Guo L, Qian Q, Gao Z (2016) QTL analysis for chalkiness of rice and fine mapping of a candidate gene for qACE9. Rice 9(1):41. https://doi.org/10.1186/s12284-016-0114-5
Chen LK, Gao WW, Chen SP, Wang LP, Zou JY, Liu YZ, Wang H, Chen ZQ, Guo T (2016) High-resolution QTL mapping for grain appearance traits and co-localization of chalkiness-associated differentially expressed candidate genes in rice. Rice 9:1–17. https://doi.org/10.1186/s12284-016-0121-6
Nguyen KL, Grondin A, Courtois B, Gantet P (2018) Next-generation sequencing accelerates crop gene discovery. Trends Plant Sci 24(3):263–274. https://doi.org/10.1016/j.tplants.2018.11.008
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74(1):174–183. https://doi.org/10.1111/tpj.12105
Zegeye WA, Zhang YX, Cao LY, Cheng SH (2018) Whole genome resequencing from bulked populations as a rapid QTL and gene identification method in rice. Int J Mol Sci 19(12):4000. https://doi.org/10.3390/ijms19124000
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303. https://doi.org/10.1101/gr.107524.110
Mansfeld BN, Grumet R (2018) QTLseqr: an R package for bulk segregant analysis with next-generation sequencing. Plant Genome. https://doi.org/10.3835/plantgenome2018.01.0006
Wang K, Li MY, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164. https://doi.org/10.1093/nar/gkq603
Meng L, Li HH, Zhang LY, Wang JK (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3(3):269–283. https://doi.org/10.1016/j.cj.2015.01.001
Peters JL, Cnudde F, Gerats T (2003) Forward genetics and map-based cloning approaches. Trends Plant Sci 8(10):484–491. https://doi.org/10.1016/j.tplants.2003.09.002
Gallavotti A, Whipple CJ (2015) Positional cloning in maize (Zea mays subsp. mays, Poaceae). App Plant Sci 3 (1). https://doi.org/10.3732/apps.1400092
Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen JE, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6(8):550–551. https://doi.org/10.1038/nmeth0809-550
Austin RS, Vidaurre D, Stamatiou G, Breit R, Provart NJ, Bonetta D, Zhang J, Fung P, Gong Y, Wang PW, McCourt P, Guttman DS (2011) Next-generation mapping of Arabidopsis genes. Plant J 67(4):715–725. https://doi.org/10.1111/j.1365-313x.2011.04619.x
Takagi H, Uemura A, Yaegashi H, Tamiru M, Abe A, Mitsuoka C, Utsushi H, Natsume S, Kanzaki H, Matsumura H, Saitoh H, Yoshida K, Cano LM, Kamoun S, Terauchi R (2013) MutMap-Gap: whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol 200(1):276–283. https://doi.org/10.1111/nph.12369
Takagi H, Tamiru M, Abe A, Yoshida K, Uemura A, Yaegashi H, Obara T, Oikawa K, Utsushi H, Kanzaki E, Mitsuoka C, Natsume S, Kosugi S, Kanzaki H, Matsumura H, Urasaki N, Kamoun S, Terauchi R (2015) MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat Biotechnol 33(5):445–449. https://doi.org/10.1038/nbt.3188
Wambugu P, Ndjiondjop MN, Furtado A, Henry R (2018) Sequencing of bulks of segregants allows dissection of genetic control of amylose content in rice. Plant Biotechnol J 16(1):100–110. https://doi.org/10.1111/pbi.12752
Arikit S, Wanchana S, Khanthong S, Saensuk C, Thianthavon T, Vanavichit A, Toojinda T (2019) QTL-seq identifies cooked grain elongation QTLs near soluble starch synthase and starch branching enzymes in rice (Oryza sativa L). Sci Rep 9(1):8328. https://doi.org/10.1038/s41598-019-44856-2
Lahari Z, Ribeiro A, Talukdar P, Martin B, Heidari Z, Gheysen G, Price AH, Shrestha R (2019) QTL-seq reveals a major root-knot nematode resistance locus on chromosome 11 in rice (Oryza sativa L). Euphytica 215(7):117. https://doi.org/10.1007/s10681-019-2427-0
Kamolsukyeunyong W, Ruengphayak S, Chumwong P, Kusumawati L, Chaichoompu E, Jamboonsri W, Saensuk C, Phoonsiri K, Toojinda T, Vanavichit A (2019) Identification of spontaneous mutation for broad-spectrum brown planthopper resistance in a large, long-term fast neutron mutagenized rice population. Rice 12(1):16. https://doi.org/10.1186/s12284-019-0274-1
Guo LA, Chen WL, Tao L, Hu BH, Qu GL, Tu B, Yuan H, Ma BT, Wang YP, Zhu XB, Qin P, Li SG (2020) GWC1 is essential for high grain quality in rice. Plant Sci 296:110497
Li Y, Fan C, Xing Y, Yun P, Luo L, Yan B, Peng B, Xie W, Wang G, Li X, Xiao J, Xu C, He Y (2014) Chalk5 encodes a vacuolar H(+)-translocating pyrophosphatase influencing grain chalkiness in rice. Nat Genet 46(4):398–404. https://doi.org/10.1038/ng.2923
Wu B, Yun P, Zhou H, Xia D, Gu Y, Li P, Yao J, Zhou Z, Chen J, Liu R, Cheng S, Zhang H, Zheng Y, Lou G, Chen P, Wan S, Zhou M, Li Y, Gao G, Zhang Q, Li X, Lian X, He Y (2022) Natural variation in WHITE-CORE RATE 1 regulates redox homeostasis in rice endosperm to affect grain quality. Plant Cell 34(5):1912–1932. https://doi.org/10.1093/plcell/koac057
Jiang L, Zhong H, Jiang X, Zhang J, Huang R, Liao F, Deng Y, Liu Q, Huang Y, Wang H, Tao Y, Zheng J (2021) Identification and pleiotropic effect analysis of GSE5 on rice chalkiness and grain shape. Front Plant Sci 12:814928. https://doi.org/10.3389/fpls.2021.814928
Wan XY, Wan JM, Weng JF, Jiang L, Bi JC, Wang CM, Zhai HQ (2005) Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor Appl Genet 110(7):1334–1346. https://doi.org/10.1007/s00122-005-1976-x
Misra G, Badoni S, Parween S, Singh RK, Leung H, Ladejobi O, Mott R, Sreenivasulu N (2021) Genome-wide association coupled gene to gene interaction studies unveil novel epistatic targets among major effect loci impacting rice grain chalkiness. Plant Biotechnol J 19(5):910–925. https://doi.org/10.1111/pbi.13516
Quero G, Gutierrez L, Monteverde E, Blanco P, Perez de Vida F, Rosas J, Fernandez S, Garaycochea S, McCouch S, Berberian N, Simondi S, Bonnecarrere V (2018) Genome-wide association study using historical breeding populations discovers genomic regions involved in high-quality rice. Plant Genome 11(3):170076. https://doi.org/10.3835/plantgenome2017.08.0076
Acknowledgements
The authors are grateful to all the members in rice group of Henan Agricultrual University.
Funding
This work was supported by the Key Research Project of Henan Higher Education (Grant No. 21A210019), Mordern Agricultural Technology System of Henan Province (Grant No. S2012-04-G02) and Central Plains Talents Program of Henan Province (Talent Training Series)—Top Young Talents in Central Plains (ZYYCYU202012170).
Author information
Authors and Affiliations
Contributions
HS and YD contributed to the study’s conception and design. Experiments was done by ZY, FL and QZ. TP and JL helped in data analysis. HS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declared that they had no conflicts of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2023_8537_MOESM1_ESM.pdf
Supplementary file1 (PDF 163 KB) Fig. S1 Chromosome distribution of polymorphic SNPs and InDels between japonica rice varieties Shuijing3 and Lamujia. The color indicated numbers of polymorphic sites in each 10kb window size.
11033_2023_8537_MOESM2_ESM.pdf
Supplementary file2 (PDF 187 KB) Fig. S2 SNP index of the BC1F2 population QTL-seq. A, SNP index of high chalkiness bulk; B, SNP index of low chalkiness bulk; C, delta SNP index between high and low chalkiness bulk.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, H., Yuan, Z., Li, F. et al. Mapping of qChalk1 controlling grain chalkiness in japonica rice. Mol Biol Rep 50, 5879–5887 (2023). https://doi.org/10.1007/s11033-023-08537-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08537-8