Abstract
Background
Breast cancer (BC) is the second leading cause of cancer-related mortality among women. Beyond the established tumourigenic role of genetic mutations, metabolic reprogramming is another key cancer hallmark. Glucose metabolism in particular is known to be prominently altered in tumours, in order to support biomass accumulation and cancer cell survival. The tumor suppressor microRNA (miRNA) miR-22 has been previously associated with a plethora of BC phenotypes such as growth, invasion-metastasis, and regulation of metabolic phenotypes such as lipid and folate metabolism. In this study, we aimed to investigate the role of miR-22 in the regulation of glucose metabolism in BC cells.
Methods and results
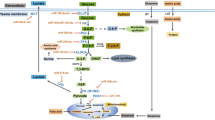
Here we examined how miR-22 affects glucose metabolism in the MCF-7 BC cells. We found that over-expression of miR-22 caused a reduced glycolytic rate in these cells. Moreover, the miRNA also rendered MCF-7 cells more sensitive to lower glucose levels. We next unbiasedly screened the transcript levels of 84 genes relevant to glucose metabolism using the Human Glucose RT2 Profiler PCR Array. Interestingly, the strongest effect identified by this screen was the upregulation of genes involved in glycogen synthesis and the repression of gene involved in glycogen catabolism. Examination of publicly available transcriptomic datasets confirmed the correlations between expression of miR-22 and these glycogen metabolism genes in BC cells.
Conclusion
This study has generated evidence for a regulatory role of miR-22 in glucose and glycogen metabolism, expanding the involvement of this miRNA in BC metabolic reprogramming.





Similar content being viewed by others
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- miRNA:
-
microRNA
- BC:
-
Breast cancer
- miR22HG:
-
miR-22 host gene
References
Bartel DP (2018) Metazoan MicroRNAs. Cell 173:20–51. https://doi.org/10.1016/j.cell.2018.03.006
Gebert LFR, MacRae IJ (2019) Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 20:21–37. https://doi.org/10.1038/s41580-018-0045-7
Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK (2016) miRNA deregulation in Cancer cells and the Tumor Microenvironment. Cancer Discov 6:235–246. https://doi.org/10.1158/2159-8290.CD-15-0893
Arnold M, Morgan E, Rumgay H et al (2022) Current and future burden of breast cancer: global statistics for 2020 and 2040. The Breast 66:15–23. https://doi.org/10.1016/j.breast.2022.08.010
Mahdavi M, Nassiri M, Kooshyar MM et al (2019) Hereditary breast cancer; genetic penetrance and current status with BRCA. J Cell Physiol 234:5741–5750. https://doi.org/10.1002/jcp.27464
Reis-Filho JS, Pusztai L (2011) Gene expression profiling in breast cancer: classification, prognostication, and prediction. The Lancet 378:1812–1823. https://doi.org/10.1016/S0140-6736(11)61539-0
Christodoulou F, Raible F, Tomer R et al (2010) Ancient animal microRNAs and the evolution of tissue identity. Nature 463:1084–1088. https://doi.org/10.1038/nature08744
Chou J, Kaller M, Jaeckel S et al (2022) AP4 suppresses DNA damage, chromosomal instability and senescence via inducing MDC1/Mediator of DNA damage checkpoint 1 and repressing MIR22HG/miR-22-3p. Mol Cancer 21:120. https://doi.org/10.1186/s12943-022-01581-1
Zhang D-Y, Zou X-J, Cao C-H et al (2018) Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for Hepatocellular Carcinoma. Theranostics 8:3751–3765. https://doi.org/10.7150/thno.22493
Ke J, Wang Q, Zhang W et al (2022) LncRNA MIR4435-2HG promotes proliferation, migration, invasion and epithelial mesenchymal transition via targeting miR-22-3p/TMEM9B in breast cancer. Am J Transl Res 14:5441–5454
Fan T, Wang C-Q, Li X-T et al (2021) MiR-22-3p suppresses Cell Migration and Invasion by Targeting PLAGL2 in breast Cancer. J Coll Physicians Surg Pak 31:937–940. https://doi.org/10.29271/jcpsp.2021.08.937
Wang X, Yao Z, Fang L (2021) miR-22-3p/PGC1β Suppresses Breast Cancer Cell Tumorigenesis via PPARγ. PPAR Res 2021:6661828. https://doi.org/10.1155/2021/6661828
Pandey DP, Picard D (2009) miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol 29:3783–3790. https://doi.org/10.1128/MCB.01875-08
Xiong J, Yu D, Wei N et al (2010) An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J 277:1684–1694. https://doi.org/10.1111/j.1742-4658.2010.07594.x
Kong L-M, Liao C-G, Zhang Y et al (2014) A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res 74:3764–3778. https://doi.org/10.1158/0008-5472.CAN-13-3555
Sun L, Liu X, Song S et al (2021) Identification of LIG1 and LIG3 as prognostic biomarkers in breast cancer. Open Med (Wars) 16:1705–1717. https://doi.org/10.1515/med-2021-0388
Patel JB, Appaiah HN, Burnett RM et al (2011) Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene 30:1290–1301. https://doi.org/10.1038/onc.2010.510
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674. https://doi.org/10.1016/j.cell.2011.02.013
WARBURG O (1956) On respiratory impairment in cancer cells. Science 124:269–270
vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033. https://doi.org/10.1126/science.1160809
Shin E, Koo JS (2021) Glucose metabolism and glucose transporters in breast Cancer. Front Cell Dev Biol 9:728759. https://doi.org/10.3389/fcell.2021.728759
Yang L, Li J, Li Y et al (2021) Diclofenac impairs the proliferation and glucose metabolism of triple-negative breast cancer cells by targeting the c-Myc pathway. Exp Ther Med 21:584. https://doi.org/10.3892/etm.2021.10016
Marini C, Salani B, Massollo M et al (2013) Direct inhibition of hexokinase activity by metformin at least partially impairs glucose metabolism and tumor growth in experimental breast cancer. Cell Cycle 12:3490–3499. https://doi.org/10.4161/cc.26461
Koufaris C, Valbuena GN, Pomyen Y et al (2016) Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene 35:2766–2776. https://doi.org/10.1038/onc.2015.333
Liu Y, Li H, Liu Y, Zhu Z (2018) MiR-22-3p targeting alpha-enolase 1 regulates the proliferation of retinoblastoma cells. Biomed Pharmacother 105:805–812. https://doi.org/10.1016/j.biopha.2018.06.038
Chen B, Tang H, Liu X et al (2015) miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett 356:410–417. https://doi.org/10.1016/j.canlet.2014.09.028
Song T-R, Su G-D, Chi Y-L et al (2021) Dysregulated miRNAs contribute to altered placental glucose metabolism in patients with gestational diabetes via targeting GLUT1 and HK2. Placenta 105:14–22. https://doi.org/10.1016/j.placenta.2021.01.015
Lone SN, Maqbool R, Parray FQ, Ul Hussain M (2018) Triose-phosphate isomerase is a novel target of miR-22 and miR-28, with implications in tumorigenesis. J Cell Physiol 233:8919–8929. https://doi.org/10.1002/jcp.26821
Jiang X, Hu C, Arnovitz S et al (2016) miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat Commun 7:11452. https://doi.org/10.1038/ncomms11452
Garofalo M, Quintavalle C, Romano G et al (2012) miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med 12:27–33. https://doi.org/10.2174/156652412798376170
Nejati K, Alivand MR, Arabzadeh AA (2021) MicroRNA-22 in female malignancies: focusing on breast, cervical, and ovarian cancers. Pathol Res Pract 223:153452. https://doi.org/10.1016/J.PRP.2021.153452
Rousset M, Zweibaum A, Fogh J (1981) Presence of glycogen and growth-related variations in 58 cultured human tumor cell lines of various tissue origins. Cancer Res 41:1165–1170
Favaro E, Bensaad K, Chong MG et al (2012) Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab 16:751–764. https://doi.org/10.1016/j.cmet.2012.10.017
Zois CE, Hendriks AM, Haider S et al (2022) Liver glycogen phosphorylase is upregulated in glioblastoma and provides a metabolic vulnerability to high dose radiation. Cell Death Dis 13:573. https://doi.org/10.1038/s41419-022-05005-2
Funding
HK, CK, and JE acknowledge support by the European Community’s Seventh Framework Programme—Health (FP7/2007–2013) project DETECTIVE (grant agreement number 266838).
Author information
Authors and Affiliations
Contributions
Conceptualization: Costas Koufaris, Hector C Keun; Methodology: Costas Koufaris, Margarita E Papandreou, James K Ellis, Vicky Nicolaidou, Hector C Keun; Formal analysis and investigation: Costas Koufaris; Writing - original draft preparation: Costas Koufaris, Vicky Nicolaidou; Writing - review and editing: Costas Koufaris, Margarita E Papandreou, James K Ellis, Vicky Nicolaidou, Hector C Keun; Funding acquisition: Hector C Keun; Resources: Hector C Keun; Supervision: Costas Koufaris, Hector C Keun.
Corresponding author
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Koufaris, C., Papandreou, M.E., Ellis, J.K. et al. miR-22-enriched breast cancer cells display repressed glycolytic metabolism, increased glycogen synthesis, and reduced survival in low glucose conditions. Mol Biol Rep 50, 5185–5193 (2023). https://doi.org/10.1007/s11033-023-08458-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08458-6