Abstract
Background
Macrobrachium nipponense, is an important economic indigenous prawn and is widely distributed in China. However, most these genetic structure analysis researches were focused on a certain water area, systematic comparative studies on genetic structure of M. nipponense across China are not yet available.
Methods and results
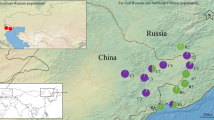
In this study, D-loop region sequences was used to investigate the genetic diversity and population structure of 22 wild populations of M. nipponense through China, containing the major rivers and lakes of China. Totally 473 valid D-loop sequences with a length of 1110 bp were obtained, and 348 variation sites and 221 haplotypes were detected. The haplotype diversity (h) was ranged from 0.1630 (Bayannur) ~ 1.0000 (Amur River) and the nucleotide diversity π value ranged from 0.001164 (Min River) ~ 0.037168 (Nen River). The pairwise genetic differentiation index (FST) ranged from 0.00344 to 0.91243 and most pair-wised FST was significant (P < 0.05). The lowest FST was displayed in Min River and Jialing River populations and the highest was between Nandu River and Nen River populations. The phylogenetic tree of genetic distance showed that all populations were divided into two branches. The Dianchi Lake, Nandu River, Jialing River and Min River populations were clustered into one branch. The neutral test and mismatch distribution results showed that M. nipponense populations were not experienced expanding and kept a steady increase.
Conclusions
Taken together, a joint resources protection and management strategy for M. nipponense have been suggested based on the results of this study for its sustainable use.





Similar content being viewed by others
References
Cai Y, Shokita S (2006) Report on a collection of freshwater shrimps (Crustacea: Decapoda: Caridea) from the Philippines, with descriptions of four new species. Raffles Bullet Zool 54(2):245–270
Fu HT, Gong YS, Wu Y, Xu P, Wu CJ (2004) Artificial interspecific hybridization between Macrobrachium species. Aquaculture 232:215–223
Fu HT, Jiang SF, Xiong YW (2012) Current status and prospects of farming the giant river prawn (Macrobrachium rosenbergii) and the oriental river prawn (Macrobrachium nipponense) in China. Aquac Res 43(7):993–998. https://doi.org/10.1111/j.1365-2109.2011.03085.x
Zhang W, Jiang SF, Salumy KR, Xuan Z, Xiong Y, Jin SB, Fu HT (2022) Comparison of genetic diversity and population structure of eight Macrobrachium nipponense populations in China based on D-loop sequences. Aquaculture Reports 23:10108. https://doi.org/10.1016/j.aqrep.2022.10108
Fu, H.T., Jin, S.B., (2018). Culture of the oriental river prawn (Macrobrachium nipponense). Aquaculture in China: Success stories and modern trends. 218–225.
Ge, J., Xu, Z., Huang, Y., Lu, Q., Pan, J., Yang, J., (2011). Genetic Variation in Wild and Cultured Populations of the Freshwater Prawn, Macrobrachium nipponense, in China. Journal of the World Aquaculture Society, 42(4), 504–511.https://doi.org/10.1111/j.1749-7345.2011.00492.x
Ma KY, Feng JB, Li JL (2012) Genetic variation based on microsatellite analysis of the oriental river prawn, Macrobrachium nipponense from Qiandao Lake in China. Genet Mol Res 11(4):4235–4244. https://doi.org/10.3724/SP.J.1141.2011.04363
Qiao, H., Lv, D., Jiang, S.F., Sun, S.M., Gong, Y.S., Xiong, Y.W., Fu, H.T., (2013). Genetic diversity analysis of oriental river prawn, Macrobrachium nipponense, in Yellow River using microsatellite marker. Genetics and Molecular Research, 12(4), 5694–5703.https://doi.org/10.4238/2013.November.18.18
Chen, P.C., Shih, C.H., Chu, T.J., Lee, Y.C., Tzeng, T.D., (2017). Phylogeography and genetic structure of the oriental river prawn Macrobrachium nipponense (Crustacea: Decapoda: Palaemonidae) in East Asia. PloS one, 12(3), e0173490. https://doi.org/10.1371/journal.pone.0173490
Cui F, Yu Y, Bao F, Wang S, Xiao MS (2018) Genetic diversity analysis of the oriental river prawn (Macrobrachium nipponense) in Huaihe River. Mitochondrial DNA Part A 29(5):737–744. https://doi.org/10.1080/24701394.2017.1350953
Li Y, Fan W, Huang Y, Huang Y, Du X, Liu Z, Zhao Y (2021) Comparison of morphology and genetic diversity between broodstock and hybrid offspring of oriental river prawn, Macrobrachium nipponense based on morphological analysis and SNP markers. Anim Genet 52(4):461–471. https://doi.org/10.1111/age.13081
Gallagher J, Finarelli JA, Jonasson JP, Carlsson J (2019) Mitochondrial D-loop DNA analyses of Norway lobster (Nephrops norvegicus) reveals genetic isolation between Atlantic and East Mediterranean populations. J Mar Biol Assoc UK 99(4):933–940. https://doi.org/10.1017/S0025315418000929
Song CY, Sun ZC, Gao TX, Song N (2020) Structure analysis of mitochondrial DNA control region sequences and its applications for the study of population genetic diversity of Acanthogobius ommaturus. Russ J Mar Biol 46(4):292–301. https://doi.org/10.1134/S1063074020040082
Rendón-Hernández JF, Pérez-Rostro CI, González-Trujillo R, Arcos-Ortega GF, Jiménez-Badillo MDL, Galindo-Cortes G (2021) Genetic diversity of the blue land crab Cardisoma guanhumi (Latreille 1825) in the Alvarado Lagoon System in Veracruz, gulf coast of Mexico. Lat Am J Aquat Res 49(1):136–147. https://doi.org/10.3856/vol49-issue1-fulltext-2478
Pan Z, Zhao H, Zhu C, Chen H, Zhao P, Cheng Y (2021) Genetic diversity analysis of crucian carp (Carassius auratus) based on cyt b and D-loop-containing region around Hongze Lake. Environ Biol Fishes 1–20:10. https://doi.org/10.1007/s10641-021-01175-8
Peng M, Zhu W, Yang C, Yao J, Chen H, Jiang W, Chen X (2021) Genetic diversity of mitochondrial D-LOOP sequences in the spotted scat (Scatophagus argus) from different geographical populations along the northern coast of the South China Sea. J Appl Ichthyol 37(1):73–82. https://doi.org/10.1111/jai.14121
Englund, T., Stromstedt, L., Johansson, A., (2014). Relatedness and diversity of nine Swedish local chicken breeds as indicated by the mtDNA D-loop. Hereditas 151 (6), 229e233. https://doi.org/10.1111/hrd2.00064
Zhang, J., Jiao, T., Zhao, S., (2016). Genetic diversity in the mitochondrial DNA D-loop region of global swine (Sus scrofa) populations. Biochem. Biophys. Res. Commun. 473 (4), 814e820. https://doi.org/10.1016/j.bbrc.2016.03.125
Jiang B, Fu J, Dong Z, Fang M, Zhu W, Wang L (2019) Maternal ancestry analyses of red tilapia strains based on D-loop sequences of seven tilapia populations. PeerJ. https://doi.org/10.7717/peerj.7007
Hall TA (1999) BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nuclc Acids Symposium Series 41:95–98. https://doi.org/10.1093/oxfordjournals.molbev.a004062
Excoffier, L., Lischer, H.E., (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular ecology resources, 10(3), 564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci 101(42):15261–15264. https://doi.org/10.1093/jhered/esab078
Beati L, Nava S, Burkman, E.J., (2013). Amblyomma cajennense (Fabricius, 1787) (Acari: Ixodidae), the Cayenne tick: phylogeography and evidence for allopatric speciation. Bmc Evolutionary Biology, 2013, 13(1): 267–267. https://doi.org/10.1186/1471-2148-13-267
Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci 86(16):6196–6200. https://doi.org/10.1676/13-054.1
Wilson K (2001) Phylogeography and systematics of the mud turtle, kinosternon baurii. Copeia 2001(3):797–801. https://doi.org/10.1643/0045-8511(2001)001[0797:PASOTM]2.0.CO;2
Wang YJ, Lu JH, Liu Z, Zhang JP (2021) Genetic diversity of Gymnocypris chilianensis (Cypriniformes, Cyprinidae) unveiled by the mitochondrial DNA D-loop region. Mitochondrial DNA Part B 6(4):1292–1297. https://doi.org/10.1080/23802359.2021.1906172
Grant WAS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered 89(5):415–426. https://doi.org/10.1093/jhered/89.5.415
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236(4803):787–792. https://doi.org/10.1016/0198-0254(87)90895-8
Sun N, Zhu DM, Li Q, Wang GY, Chen J, Zheng F, Sun YH (2021) Genetic diversity analysis of Topmouth Culter (Culter alburnus) based on microsatellites and D-loop sequences. Environ Biol Fishes 104(3):213–228. https://doi.org/10.1007/s10641-021-01062-2
Gao W, Zeng Y, Zhao D, Wu B, Ren Z (2020) Land cover changes and drivers in the water source area of the Middle Route of the South-to-North Water Diversion Project in China from 2000 to 2015. Chin Geogra Sci 30(1):115–126. https://doi.org/10.1007/s11769-020-1099-y
Walker K, Yang H (1999) Fish and fisheries in western China. Fish Fish Higher Altitudes: Asia 385:304
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147(2):915–925. https://doi.org/10.1093/genetics/147.2.915
De Oliveira-Neto JF, Pie MR, Chammas MA, Ostrensky A, Boeger WA (2008) Phylogeography of the blue land crab, Cardisoma guanhumi (Decapoda Gecarcinidae) along the Brazilian coast. J Mar Biol Ass United Kingdom 88(7):1417–1423
Neraas LP, Spruell P (2001) Fragmentation of riverine systems: the genetic effects of dams on bull trout (Salvelinus confluentus) in the Clark Fork River system. Mol Ecol 10(5):1153–1164. https://doi.org/10.1046/j.1365-294X.2001.01269.x
Cook DR, Sullivan SMP (2018) Associations between riffle development and aquatic biota following lowhead dam removal. Environ Monit Assess 190(6):1–14. https://doi.org/10.1007/s1061-081-6716-1
Funding
This research was supported by grants from Central Public-interest Scientific Institution Basal Research Fund CAFS (2020TD36); Jiangsu Agricultural Industry Technology System; the National Key R&D Program of China (2018YFD0901303); the earmarked fund for CARS-48; the New cultivar breeding Major Project of Jiangsu province (PZCZ201745). The seed industry revitalization project of Jiangsu province (JBGS [2021] 118). Thanks for the Jiangsu Province Platform for the Conservation and Utilization of Agricultural Germplasm.
Author information
Authors and Affiliations
Contributions
Hongtuo Fu and Hui Qiao designed the research; Sufei Jiang, Yiwei, Xiong collect samples and performed the study; Lijuan Zhang and Wenyi Zhang analyzed the data;, Jisheng Wang, Yalu Zheng, Shubo Jin, Yongsheng Gong and Yan Wu contributed reagents, materials and tools; Sufei Jiang and Hui Qiao drafted the manuscript; Hongtuo Fu revised the manuscript; all authors approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Ethical approval
No endangered or protected species were involved in this study and all experimental protocols, methods were approved in July 2021 (Authorization NO. 20211105001) by the Animal Care and Use Ethics Committee in the Freshwater Fisheries Research Center (Wuxi, China).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to publish
The authors affirm that human research participants provided informed consent for publication of the images in Figure(s) 1, 2,3 and 4.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, S., Xiong, Y., Zhang, L. et al. Genetic diversity and population structure of wild Macrobrachium nipponense populations across China: Implication for population management. Mol Biol Rep 50, 5069–5080 (2023). https://doi.org/10.1007/s11033-023-08402-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08402-8