Abstract
Background
Synonymous variations have always been ignored while studying the underlying genetic mechanisms for most of the human diseases. However, recent studies have suggested that these silent changes in the genome can alter the protein expression and folding.
Methods and results
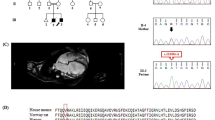
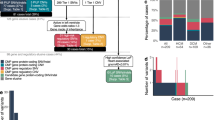
CSRP3, which is a well-known candidate gene associated with dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM), was screened for 100 idiopathic DCM cases and 100 controls. Three synonymous variations were identified viz., c.96G > A, p.K32=; c.336G > A, p.A112=; c.354G > A, p.E118=. A comprehensive in silico analysis was performed using various web based widely accepted tools, Mfold, Codon Usage, HSF3.1 and RNA22. Mfold predicted structural changes in all the variants except c.96 G > A (p.K32=), however it predicted changes in the stability of mRNA due to all the synonymous variants. Codon bias was observed as evident by the Relative Synonymous Codon Usage and Log Ratio of Codon Usage Frequencies. The Human Splicing Finder also predicted remarkable changes in the regulatory elements in the variants c.336G > A and c.354 G > A. The miRNA target prediction using varied modes available in RNA22 revealed that 70.6% of the target sites of miRNAs in CSRP3 were altered due to variant c.336G > A while 29.41% sites were completely lost.
Conclusion
Findings of the present study suggest that synonymous variants revealed striking deviations in the structural conformation of mRNA, stability of mRNA, relative synonymous codon usage, splicing and miRNA binding sites from the wild type suggesting their possible role in the pathogenesis of DCM, either by destabilizing the mRNA structure, or codon usage bias or else altering the cis-acting regulatory elements during splicing.



Similar content being viewed by others
Data Availability
The raw or derived data associated with the present study are available from corresponding author on request.
Code Availability
Not Applicable.
References
McKenna WJ, Maron BJ, Thiene G (2017) Classification, epidemiology, and global burden of cardiomyopathies. Circul Res 121(7):722–730. https://doi.org/10.1161/CIRCRESAHA.117.309711
Giri P, Mukhopadhyay A, Gupta M, Mohapatra B (2021) Dilated cardiomyopathy: a new insight into the rare but common cause of heart failure. Heart Fail Rev 10:1–24. https://doi.org/10.1007/s10741-021-10125-6
Komar AA, Lesnik T, Reiss C (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 462(3):387–391. https://doi.org/10.1016/S0014-5793(99)01566-5
Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 4285–298. https://doi.org/10.1038/nrg775
Plotkin JB, Kudla G (2011) Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 132–42. https://doi.org/10.1038/nrg2899
Zolk O, Caroni P, Böhm M (2000) Decreased expression of the cardiac LIM domain protein MLP in chronic human heart failure. Circulation 101(23):2674–2677. https://doi.org/10.1161/01.CIR.101.23.2674
Knöll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W (2002) The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111(7):943–955. https://doi.org/10.1016/S0092-8674(02)01226-6
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415. https://doi.org/10.1093/nar/gkg595
Bennetzen JL, Hall BD (1982) Codon selection in yeast. J Biol Chem 257(6):3026–3031. https://doi.org/10.1016/S0021-9258(19)81068-2
Mirsafian H, Mat Ripen A, Singh A, Teo PH, Merican AF, Mohamad SB (2014) A comparative analysis of synonymous codon usage bias pattern in human albumin superfamily. The Scientific World Journal 2014
Sharp PM, Li WH (1987) The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15(3):1281–1295. https://doi.org/10.1093/nar/15.3.1281
Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126(6):1203–1217. https://doi.org/10.1016/j.cell.2006.07.031
Pagani F, Raponi M, Baralle FE (2005) Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proceedings of the National Academy of Sciences 102(18):6368-72. https://doi.org/10.1073/pnas.0502288102
Pechmann S, Frydman J (2013) Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20(2):237. https://doi.org/10.1038/nsmb.2466
Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, Coller J (2015) Codon optimality is a major determinant of mRNA stability. Cell 160(6):1111–1124. https://doi.org/10.1016/j.cell.2015.02.029
Stergachis AB, Haugen E, Shafer A, Fu W, Vernot B, Reynolds A, Raubitschek A, Ziegler S, LeProust EM, Akey JM, Stamatoyannopoulos JA (2013) Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342(6164):1367–1372. https://doi.org/10.1126/science.1243490
Angellotti MC, Bhuiyan SB, Chen G, Wan XF (2007) CodonO: codon usage bias analysis within and across genomes. Nucleic Acids Res 35(suppl2):W132–W136. https://doi.org/10.1093/nar/gkm392
Dixit R, Kumar A, Mohapatra B (2019) Implication of GATA4 synonymous variants in congenital heart disease: a comprehensive in-silico approach. Mutat Research/Fundamental Mol Mech Mutagen 813. https://doi.org/10.1016/j.mrfmmm.2018.12.002. :31 – 8
Yadav ML, Jain D, Agrawal D, Kumar A, Mohapatra B (2021) A gain-of-function mutation in CITED2 is associated with congenital heart disease. Mutat Research/Fundamental Mol Mech Mutagen 822:111741. https://doi.org/10.1016/j.mrfmmm.2021.111741
Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463:457–463. https://doi.org/10.1038/nature08909
Adamson SI, Zhan L, Graveley BR (2018) Vex-seq: high-throughput identification of the impact of genetic variation on pre-mRNA splicing efficiency. Genome Biol 19(1):1–2. https://doi.org/10.1186/s13059-018-1437-x
Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB (2004) RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res 32(suppl2):W187–W190. https://doi.org/10.1093/nar/gkh393
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31(13):3568–3571. https://doi.org/10.1093/nar/gkg616
Zhang XH, Chasin LA (2004) Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 18:1241–1250. https://doi.org/10.1101/gad.1195304
Zhang C, Li WH, Krainer AR, Zhang MQ (2008) RNA landscape of evolution for optimal exon and intron discrimination. Proc. Natl Acad. Sci. USA105:5797–5802. https://doi.org/10.1073/pnas.0801692105
Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, Giorda R, Pozzoli U (2004) Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res 32:1783–1791. https://doi.org/10.1093/nar/gkh341
Arber S, Halder G, Caroni P (1994) Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell 79(2):221–231. https://doi.org/10.1016/0092-8674(94)90192-9
Gehmlich K, Geier C, Osterziel KJ, Van der Ven PF, Fürst DO (2004) Decreased interactions of mutant muscle LIM protein (MLP) with N-RAP and α-actinin and their implication for hypertrophic cardiomyopathy. Cell Tissue Res 317(2):129–136. https://doi.org/10.1007/s00441-004-0873-y
Mohapatra B, Jimenez S, Lin JH, Bowles KR, Coveler KJ, Marx JG, Chrisco MA, Murphy RT, Lurie PR, Schwartz RJ, Elliott PM (2003) Mutations in the muscle LIM protein and α-actinin-2 genes in dilated cardiomyopathy and endocardialfibroelastosis. Mol Genet Metab 80(1–2):207–215. https://doi.org/10.1016/S1096-7192(03)00142-2
Bos JM, Poley RN, Ny M, Tester DJ, Xu X, Vatta M, Towbin JA, Gersh BJ, Ommen SR, Ackerman MJ (2006) Genotype–phenotype relationships involving hypertrophic cardiomyopathy-associated mutations in titin, muscle LIM protein, and telethonin. Mol Genet Metab 88(1):78–85. https://doi.org/10.1016/j.ymgme.2005.10.008
Zimmerman RS, Cox S, Lakdawala NK, Cirino A, Mancini-DiNardo D, Clark E, Leon A, Duffy E, White E, Baxter S, Alaamery M (2010) A novel custom resequencing array for dilated cardiomyopathy. Genet Sci 12(5):268–278. https://doi.org/10.1097/GIM.0b013e3181d6f7c0
Janin A, Bessière F, Chauveau S, Chevalier P, Millat G (2018) First identification of homozygous truncating CSRP3 variants in two unrelated cases with hypertrophic cardiomyopathy. Gene 676:110–116. https://doi.org/10.1016/j.gene.2018.07.036
Lipari M, Wypasek E, Karpiński M, Tomkiewicz-Pajak L, Laino L, Binni F, Giannarelli D, Rubiś P, Petkow-Dimitrow P, Undas A, Grammatico P (2020) Identification of a variant hotspot in MYBPC3 and of a novel CSRP3 autosomal recessive alteration in a cohort of polish patients with hypertrophic cardiomyopathy. Pol archives Intern Med 130(2):89–99. https://doi.org/10.20452/pamw.15130
Diederichs S, Bartsch L, Berkmann JC, Fröse K, Heitmann J, Hoppe C, Iggena D, Jazmati D, Karschnia P, Linsenmeier M, Maulhardt T (2016) The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med 8(5):442–457. https://doi.org/10.15252/emmm.201506055
Wen P, Xiao P, Xia J (2016) dbDSM: a manually curated database for deleterious synonymous mutations. Bioinformatics 32(12):1914–1916. https://doi.org/10.1093/bioinformatics/btw086
Sauna ZE, Kimchi-Sarfaty C (2011) Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12(10):683–691. https://doi.org/10.1038/nrg3051
Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B (2014) Synonymous mutations frequently act as driver mutations in human cancers. Cell 156(6):1324–1335. https://doi.org/10.1016/j.cell.2014.01.051
Hunt RC, Simhadri VL, Iandoli M, Sauna ZE, Kimchi-Sarfaty C (2014) Exposing synonymous mutations. Trends Genet 30(7):308–321. https://doi.org/10.1016/j.tig.2014.04.006
Spencer PS, Siller E, Anderson JF, Barral JM (2012) Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J Mol Biol 422(3):328–335. https://doi.org/10.1016/j.jmb.2012.06.010
Edwards NC, Hing ZA, Perry A, Blaisdell A, Kopelman DB, Fathke R, Plum W, Newell J, Allen CE, Shapiro A, Okunji C (2012) Characterization of coding synonymous and non-synonymous variants in ADAMTS13 using ex vivo and in silico approaches. PLoS ONE 7(6):e38864. https://doi.org/10.1371/journal.pone.0038864
Bertalovitz AC, Badhey ML, McDonald TV (2018) Synonymous nucleotide modification of the KCNH2 gene affects both mRNA characteristics and translation of the encoded hERG ion channel. J Biol Chem 293(31):12120–12136. https://doi.org/10.1074/jbc.RA118.001805
Sharma Y, Miladi M, Dukare S, Boulay K, Caudron-Herger M, Groß M, Backofen R, Diederichs S (2019) A pan-cancer analysis of synonymous mutations. Nat Commun 10(1):1–4. https://doi.org/10.1038/s41467-019-10489-2
Gaspar P, Moura G, Santos MA, Oliveira JL (2013) mRNA secondary structure optimization using a correlated stem–loop prediction. Nucleic Acids Res 41(6):e73–e73
de Smit MH, van Duin J (1994) Control of translation by mRNA secondary structure in Escherichia coli: a quantitative analysis of literature data. J Mol Biol 244(2):144–150
Hall MN, Gabay J, Débarbouillé M, Schwartz M (1982) A role for mRNA secondary structure in the control of translation initiation. Nature 295(5850):616–618
Kozak M (1986) Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proceedings of the National Academy of Sciences, 83(9), 2850–2854
Studer SM, Joseph S (2006) Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell 22(1):105–115
Faure G, Ogurtsov AY, Shabalina SA, Koonin EV (2016) Role of mRNA structure in the control of protein folding. Nucleic Acids Res 44(22):10898–10911
Wen, J. D., Lancaster, L., Hodges, C., Zeri, A. C., Yoshimura, S. H., Noller, H. F.,… Tinoco, I. (2008). Following translation by single ribosomes one codon at a time.Nature, 452(7187), 598–603
Locker N, Chamond N, Sargueil B (2011) A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res 39(6):2367–2377
Watts, J. M., Dang, K. K., Gorelick, R. J., Leonard, C. W., Bess Jr, J. W., Swanstrom,R., … Weeks, K. M. (2009). Architecture and secondary structure of an entire HIV-1 RNA genome. Nature, 460(7256), 711–716
Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM (2007) A” silent” polymorphism in the MDR 1 gene changes substrate specificity. Science 315(5811):525–528
Fung KL, Hunt RC, Kimchi-Sarfaty C, Gottesman MM (2016) Genetic polymorphisms of P-glycoprotein: echoes of silence. ABC Transporters-40 years on. Springer, Cham, pp 105–134
Bartoszewski, R. A., Jablonsky, M., Bartoszewska, S., Stevenson, L., Dai, Q., Kappes,J., … Bebok, Z. (2010). A synonymous single nucleotide polymorphism in ∆F508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. Journal of Biological Chemistry, 285(37), 28741–28748
Ge WW, Leystra-Lantz C, Sanelli TR, McLean J, Wen W, Strong W, Strong MJ (2006) Neuronal tissue-specific ribonucleoprotein complex formation on SOD1 mRNA: alterations by ALS SOD1 mutations. Neurobiol Dis 23(2):342–350
Shabalina SA, Spiridonov NA, Kashina A (2013) Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res 41(4):2073–2094
Zhang F, Saha S, Shabalina SA, Kashina A (2010) Differential arginylation of actin isoforms is regulated by coding sequence–dependent degradation. Science 329(5998):1534–1537
Zhang, F., Patel, D. M., Colavita, K., Rodionova, I., Buckley, B., Scott, D. A., …Kashina, A. (2015). Arginylation regulates purine nucleotide biosynthesis by enhancing the activity of phosphoribosyl pyrophosphate synthase. Nature communications, 6(1),1–9
Shatoff E, Bundschuh R (2020) Single nucleotide polymorphisms affect RNA-protein interactions at a distance through modulation of RNA secondary structures.PLoS Computational Biology, 16(5), e1007852
Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, Link K, Khatwani N, Reynders J, Moore MJ, McFadyen IJ (2019) mRNA structure regulates protein expression through changes in functional half-life. Proceedings of the National Academy of Sciences 116(48):24075-83. https://doi.org/10.1073/pnas.1908052116
Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM (2014) In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505(7485):696–700. https://doi.org/10.1038/nature12756
Koutmou KS, Radhakrishnan A, Green R (2015) Synthesis at the speed of codons. Trends Biochem Sci 40(12):717–718. https://doi.org/10.1016/j.tibs.2015.10.005
Mitra S, Ray SK, Banerjee R (2016) Synonymous codons influencing gene expression in organisms. Rese Rep Biochem 6:57–65. https://doi.org/10.2147/RRBC.S83483
Scotti MM, Swanson MS (2016) RNA mis-splicing in disease. Nat Rev Genet 17(1):19–32. https://doi.org/10.1038/nrg.2015.3
Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, … Chasin LA (2011) Quantitative evaluation of all hexamers as exonic splicing elements. Genome research 21(8):1360–1374 https://doi.org/10.1101/gr.119628.110
Pengelly RJ, Bakhtiar D, Borovská I, Královičová J, Vořechovský I (2022) Exonic splicing code and protein binding sites for calcium. Nucleic Acids Res 50(10):5493–5512. https://doi.org/10.1093/nar/gkac270
Latronico MV, Catalucci D, Condorelli G (2007) Emerging role of microRNAs in cardiovascular biology. Circul Res 101(12):1225–1236. https://doi.org/10.1161/CIRCRESAHA.107.163147
Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X, Harel-Bellan A (2011) A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet 43(3):242–245. https://doi.org/10.1038/ng.762
Sousa MA, de Athayde FR, Maldonado MB, Lima AO, Fortes MR, Lopes FL (2021) Single nucleotide polymorphisms affect miRNA target prediction in bovine. PLoS ONE 16(4):e0249406. https://doi.org/10.1371/journal.pone.0249406
Acknowledgements
We are grateful to all the patients, their families and control individuals who participated in the present study. We are extremely thankful to Dr. T. K. Lahiri and Dr. Damyanti Agarwal from Department of Cardio-vascular and Thoracic Surgery, IMS, BHU, Varanasi for their constant support and encouragement in recruitment of patients. We acknowledge University Grants Commission–Centre of Advanced Studies (UGC-CAS) for research fellowship to Ms. Prerna Giri.
Funding
This study was financially supported by the Department of Atomic Energy, Board of Research in Nuclear Sciences, Mumbai, India (BRNS No. 2013/37B/28). The funding agency had no role in experimental design, sample collection, analysis and interpretation of data, manuscript preparation and decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
Prerna Giri performed the mutation screening, all the in silico characterization and prepared the original draft including all the figures and table. Dr. Bhagyalaxmi Mohapatra conceptualized the study, acquired funding, critically supervised and revised the manuscript at every single step. Dr. Ashok Kumar and Dr. Dharmendra Jain clinically evaluated and diagnosed patients recruited for the study and also helped in collection of blood samples from patients and their available relatives. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of Interest/Competing interests
On behalf of all the authors, the corresponding author states that there is no conflict of interest. All the authors have read the manuscript and approved the submission of current version of the manuscript.
Financial and non-Financial interest
Authors have no relevant financial or non-financial interests to disclose.
Ethics approval
The study was approved by the ‘Institutional human ethics committee’.
Consent to participate
All the study subjects gave prior written informed consent for collection of samples.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Giri, P., Jain, D., Kumar, A. et al. Identification and in silico characterization of CSRP3 synonymous variants in dilated cardiomyopathy. Mol Biol Rep 50, 4105–4117 (2023). https://doi.org/10.1007/s11033-023-08314-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08314-7