Abstract
Background
Shark species are overfished at a global scale, as they are poached for the finning industry or are caught as bycatch. Efficient conservation measures require fine-scale spatial and temporal studies to characterize shark habitat use, infer migratory habits, analyze relatedness, and detect population genetic differentiation. Gathering these types of data is costly and time-consuming, especially when it requires collection of shark tissue samples.
Methods and results
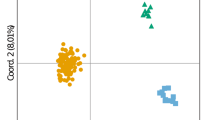
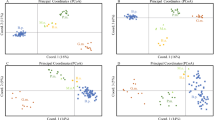
Genetic tools, such as microsatellite markers, are the most economical sampling method for collecting genetic data, as they enable the estimation of genetic diversity, population structure and parentage relationships and are thus an efficient way to inform conservation strategies. Here, a set of 45 microsatellite loci was tested on three blacktip reef shark (Carcharhinus melanopterus) populations from three Polynesian islands: Moorea, Morane and Tenararo. The set was composed of 10 previously published microsatellite markers and 35 microsatellite markers that were developed specifically for C. melanopterus as part of the present study. The 35 novel and 10 existing loci were cross-amplified on eight additional shark species (Carcharhinus amblyrhynchos, C. longimanus, C. sorrah, Galeocerdo cuvier, Negaprion acutidens, Prionacea glauca, Rhincodon typus and Sphyrna lewini). These species had an average of 69% of successful amplification, considered if at least 50% of the individual samples being successfully amplified per species and per locus.
Conclusions
This novel microsatellite marker set will help address numerous knowledge gaps that remain, concerning genetic stock identification, shark behavior and reproduction via parentage analysis.

Similar content being viewed by others
References
Alves LMF, Nunes M, Marchand P, le Bizec B, Mendes S, Correia JPS, Lemos MFL, Novais SC (2016) Blue sharks (Prionace glauca) as bioindicators of pollution and health in the Atlantic Ocean: contamination levels and biochemical stress responses. Sci Total Environ 563–564:282–292. https://doi.org/10.1016/j.scitotenv.2016.04.085
Archer FI, Adams PE, Schneiders BB (2017) StrataG: An R package for manipulating, summarizing and analyzing population genetic data. Mol Ecol Resour 17(1):5–11. https://doi.org/10.1111/1755-0998.12559
Baird SJE (2015) Exploring linkage disequilibrium. Mol Ecol Resour 15(5):1017–1019. https://doi.org/10.1111/1755-0998.12424
Balaresque P (2007) Microsatellites: life cycle concept and neutrality issues. Medicine/Sciences 23(8–9):729–734. https://doi.org/10.1051/medsci/20072389729
Barbará T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C (2007) Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol 16(18):3759–3767. https://doi.org/10.1111/j.1365-294X.2007.03439.x
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996) GENETIX 4.05, logiciel sous WindowsTM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France)
Boussarie G, Momigliano P, Robbins WD, Cornu LB, Fauvelot J-F, Kiszka C, Manel S, Mouillot D, Vigliola L (2021) Identifying barriers to gene flow and hierarchical conservation units from seascape genomics: a modelling framework applied to a marine predator. Ecography. https://doi.org/10.1101/2021.10.25.465682
Cardeñosa D, Shea SK, Zhang H, Fischer GA, Simpfendorfer CA, Chapman DD (2022) Two-thirds of species in a global shark fin trade hub are threatened with extinction: conservation potential of international trade regulations for coastal sharks. Conserv Lett. https://doi.org/10.1111/conl.12910
Chaix G, Gerber S, Razafimaharo V, Vigneron P, Verhaegen D, Hamon S (2003) Gene flow estimation with microsatellites in a Malagasy seed orchard of Eucalyptus grandis. Theor Appl Genet 107(4):705–712. https://doi.org/10.1007/s00122-003-1294-0
Clarke SC, Harley SJ, Hoyle SD, Rice JS (2013) Population trends in Pacific oceanic sharks and the utility of regulations on shark finning. Conserv Biol 27(1):197–209. https://doi.org/10.1111/j.1523-1739.2012.01943.x
Dapp DR, Huveneers C, Walker TI, Reina RD (2017) Physiological response and immediate mortality of gill-net-caught blacktip reef sharks (Carcharhinus melanopterus). Mar Freshw Res 68(9):1734. https://doi.org/10.1071/MF16132
DeWoody JA, Avise JC (2000) Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J Fish Biol 56(3):461–473. https://doi.org/10.1111/j.1095-8649.2000.tb00748.x
Domingues RR, Hilsdorf AWS, Gadig OBF (2018) The importance of considering genetic diversity in shark and ray conservation policies. Conserv Genet 19(3):501–525. https://doi.org/10.1007/s10592-017-1038-3
Dudgeon CL, Blower DC, Broderick D, Giles JL, Holmes BJ, Kashiwagi T, Krück NC, Morgan JAT, Tillett BJ, Ovenden JR (2012) A review of the application of molecular genetics for fisheries management and conservation of sharks and rays. J Fish Biol 80(5):1789–1843. https://doi.org/10.1111/j.1095-8649.2012.03265.x
Dulvy NK, Pacoureau N, Rigby CL, Pollom RA, Jabado RW, Ebert DA, Finucci B, Pollock CM, Cheok J, Derrick DH, Herman KB, Sherman CS, VanderWright WJ, Lawson JM, Walls RHL, Carlson JK, Charvet P, Bineesh KK, Fernando D et al (2021) Overfishing drives over one-third of all sharks and rays toward a global extinction crisis. Curr Biol 31(21):4773-4787.e8. https://doi.org/10.1016/j.cub.2021.08.062
Dwyer RG, Krueck NC, Udyawer V, Heupel MR, Chapman D, Pratt HL, Garla R, Simpfendorfer CA (2020) Individual and population benefits of marine reserves for reef sharks. Curr Biol. https://doi.org/10.1016/j.cub.2019.12.005
Feldheim KA, Gruber SH, Ashley MV (2002) Population genetic structure of the lemon shark (Negaprion brevirostris) in the western Atlantic: DNA microsatellite variation. Mol Ecol 10(2):295–303. https://doi.org/10.1046/j.1365-294X.2001.01182.x
Feldheim KA, Gruber SH, Ashley MV (2004) Reconstruction of parental microsatellite genotypes reveals female polyandry and philopatry in the lemon shark, Negaprion brevirostris. Evolution 58(10):2332–2342. https://doi.org/10.1111/j.0014-3820.2004.tb01607.x
Feldheim KA, Jabado RW, Chapman DD, Cardeñosa D, Maddox JD (2020) Microsatellite primer development in elasmobranchs using next generation sequencing of enriched libraries. Mol Biol Rep 47(4):2669–2675. https://doi.org/10.1007/s11033-020-05357-y
Flanagan SP, Jones AG (2019) The future of parentage analysis: from microsatellites to SNPs and beyond. Mol Ecol 28(3):544–567. https://doi.org/10.1111/mec.14988
Frichot E, François O (2015) LEA: an R package for landscape and ecological association studies. Methods Ecol Evol 6(8):925–929. https://doi.org/10.1111/2041-210X.12382
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10(2):305–318
Graham NAJ, Spalding MD, Sheppard CRC (2010) Reef shark declines in remote atolls highlight the need for multi-faceted conservation action. Aquat Conserv Mar Freshwat Ecosyst 20(5):543–548. https://doi.org/10.1002/aqc.1116
Karl SA, Castro ALF, Garla RC (2012) Population genetics of the nurse shark (Ginglymostoma cirratum) in the western Atlantic. Mar Biol 159(3):489–498. https://doi.org/10.1007/s00227-011-1828-y
Keeney DB, Heist EJ (2003) Characterization of microsatellite loci isolated from the blacktip shark and their utility in requiem and hammerhead sharks. Mol Ecol Notes 3(4):501–504. https://doi.org/10.1046/j.1471-8286.2003.00492.x
Larson SE, Daly-Engel TS, Phillips NM (2017) Review of current conservation genetic analyses of Northeast Pacific sharks. Adv Mar Biol 77:79–110. https://doi.org/10.1016/bs.amb.2017.06.005
Lei Y, Zhou Y, Price M, Song Z (2021) Genome-wide characterization of microsatellite DNA in fishes: survey and analysis of their abundance and frequency in genome-specific regions. BMC Genomics 22(1):421. https://doi.org/10.1186/s12864-021-07752-6
Li Y-C, Korol AB, Fahima T, Beiles A, Nevo E (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11(12):2453–2465. https://doi.org/10.1046/j.1365-294X.2002.01643.x
Maduna SN, Rossouw C, Roodt-Wilding R, van der Merwe AEB (2014) Microsatellite cross-species amplification and utility in southern African elasmobranchs: a valuable resource for fisheries management and conservation. BMC Res Notes. https://doi.org/10.1186/1756-0500-7-352
Martin AP, Pardini AT, Noble LR, Jones CS (2002) Conservation of a dinucleotide simple sequence repeat locus in sharks. Mol Phylogenet Evol 23(2):205–213. https://doi.org/10.1016/S1055-7903(02)00001-5
Meglécz E, Pech N, Gilles A, Dubut V, Hingamp P, Trilles A, Grenier R, Martin J-F (2014) QDD version 3.1: a user-friendly computer program for microsatellite selection and primer design revisited: experimental validation of variables determining genotyping success rate. Mol Ecol Resour 14(6):1302–1313. https://doi.org/10.1111/1755-0998.12271
Meirmans PG (2020) GENODIVE version 3.0: Easy to use software for the analysis of genetic data of diploids and polyploids. Mol Ecol Resour 20(4):1126–1131. https://doi.org/10.1111/1755-0998.13145
Mendes NJ, Cruz VP, Ashikaga FY, Camargo SM, Oliveira C, Piercy AN, Burgess GH, Coelho R, Santos MN, Mendonça FF, Foresti F (2016) Microsatellite loci in the tiger shark and cross-species amplification using pyrosequencing technology. PeerJ 4:e2205. https://doi.org/10.7717/peerj.2205
Meyer L, Fox A, Huveneers C (2018) Simple biopsy modification to collect muscle samples from free-swimming sharks. Biol Conserv 228:142–147. https://doi.org/10.1016/j.biocon.2018.10.024
Momigliano P, Harcourt R, Robbins WD, Stow A (2015) Connectivity in grey reef sharks (Carcharhinus amblyrhynchos) determined using empirical and simulated genetic data. Sci Rep 5(1):13229. https://doi.org/10.1038/srep13229
Moore JA, Draheim HM, Etter D, Winterstein S, Scribner KT (2014) Application of large-scale parentage analysis for investigating natal dispersal in highly vagile vertebrates: a case study of American black bears (Ursus americanus). PLoS ONE 9(3):e91168. https://doi.org/10.1371/journal.pone.0091168
Mourier J, Planes S (2013) Direct genetic evidence for reproductive philopatry and associated fine-scale migrations in female blacktip reef sharks (Carcharhinus melanopterus) in French Polynesia. Mol Ecol 22(1):201–214. https://doi.org/10.1111/mec.12103
Peakall R, Smouse PE (2006) GenAlex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pirog A, Jaquemet S, Blaison A, Soria M, Magalon H (2016) Isolation and characterization of eight microsatellite loci from Galeocerdo cuvier (tiger shark) and cross-amplification in Carcharhinus leucas, Carcharhinus brevipinna, Carcharhinus plumbeus and Sphyrna lewini. PeerJ 4:e2041. https://doi.org/10.7717/peerj.2041
Portnoy DS, Heist EJ (2012) Molecular markers: progress and prospects for understanding reproductive ecology in elasmobranchs. J Fish Biol 80(5):1120–1140. https://doi.org/10.1111/j.1095-8649.2011.03206.x
Portnoy DS, McDowell JR, Thompson K, Musick JA, Graves JE (2006) Isolation and characterization of five dinucleotide microsatellite loci in the sandbar shark, Carcharhinus plumbeus. Mol Ecol Notes 6(2):431–433. https://doi.org/10.1111/j.1471-8286.2006.01261.x
Ramírez-Macías D, Shaw K, Ward R, Galván-Magaña F, Vázquez-Juárez R (2009) Isolation and characterization of microsatellite loci in the whale shark (Rhincodon typus). Mol Ecol Resour 9(3):798–800. https://doi.org/10.1111/j.1755-0998.2008.02197.x
Richards VP, Suzuki H, Stanhope MJ, Shivji MS (2013) Characterization of the heart transcriptome of the white shark (Carcharodon carcharias). BMC Genomics 14(1):697. https://doi.org/10.1186/1471-2164-14-697
Roff G, Doropoulos C, Rogers A, Bozec Y-M, Krueck NC, Aurellado E, Priest M, Birrell C, Mumby PJ (2016) The ecological role of sharks on coral reefs. Trends Ecol Evol 31(5):395–407. https://doi.org/10.1016/j.tree.2016.02.014
Roncallo PF, Beaufort V, Larsen AO, Dreisigacker S, Echenique V (2019) Genetic diversity and linkage disequilibrium using SNP (KASP) and AFLP markers in a worldwide durum wheat (Triticum turgidum L. var. durum) collection. PLoS ONE 14(6):0218562. https://doi.org/10.1371/journal.pone.0218562
Salles OC, Pujol B, Maynard JA, Almany GR, Berumen ML, Jones GP, Saenz-Agudelo P, Srinivasan M, Thorrold SR, Planes S (2016) First genealogy for a wild marine fish population reveals multigenerational philopatry. Proc Natl Acad Sci USA 113(46):13245–13250. https://doi.org/10.1073/pnas.1611797113
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011(27):863–864. https://doi.org/10.1093/bioinformatics/btr026
Scotti I, Magni F, Paglia G, Morgante M (2002) Trinucleotide microsatellites in Norway spruce (Picea abies): their features and the development of molecular markers. Theor Appl Genet 106(1):40–50. https://doi.org/10.1007/s00122-002-0986-1
Sharma SP, Ghazi MG, Katdare S, Dasgupta N, Mondol S, Gupta SK, Hussain SA (2021) Microsatellite analysis reveals low genetic diversity in managed populations of the critically endangered gharial (Gavialis gangeticus) in India. Sci Rep. https://doi.org/10.1038/s41598-021-85201-w
Simpfendorfer C, Yuneni RR, Tanay D, Seyha L, Haque AB, Fahmi, Bin Ali AD, Bineesh KK, Gautama DA, Maung A, Sianipar A, Utzurrum JAT, Voye VQ (2020) Carcharhinus melanopterus. The IUCN Red List of Threatened Species. https://doi.org/10.2305/IUCN.UK.2020-3.RLTS.T39375A58303674.en
Staden M, Gledhill KS, Gennari E, McCord ME, Parkinson M, Watson RGA, Rhode C, Bester-van der Merwe AE (2020) Microsatellite development and detection of admixture among three sympatric Haploblepharus species (Carcharhiniformes: Scyliorhinidae). Aquat Conserv Mar Freshwat Ecosyst 30(12):2336–2350. https://doi.org/10.1002/aqc.3406
Sturm AB, Eckert RJ, Méndez JG, González-Díaz P, Voss JD (2020) Population genetic structure of the great star coral, Montastraea cavernosa, across the Cuban archipelago with comparisons between microsatellite and SNP markers. Sci Rep 10(1):15432. https://doi.org/10.1038/s41598-020-72112-5
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4(3):535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Vélez-Zuazo X, Agnarsson I (2011) Shark tales: a molecular species-level phylogeny of sharks (Selachimorpha, Chondrichthyes). Mol Phylogenet Evol 58(2):207–217. https://doi.org/10.1016/j.ympev.2010.11.018
Vignaud TM, Mourier J, Maynard JA, Leblois R, Spaet JLY, Clua E, Neglia V, Planes S (2014) Blacktip reef sharks, Carcharhinus melanopterus, have high genetic structure and varying demographic histories in their Indo-Pacific range. Mol Ecol 23(21):5193–5207. https://doi.org/10.1111/mec.12936
Ward-Paige CA, Worm B (2017) Global evaluation of shark sanctuaries. Glob Environ Chang. https://doi.org/10.1016/j.gloenvcha.2017.09.005
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358. https://doi.org/10.2307/2408641
Xu Y, Li W, Hu Z, Zeng T, Shen Y, Liu S, Zhang X, Li J, Yue B (2018) Genome-wide mining of perfect microsatellites and tetranucleotide orthologous microsatellites estimates in six primate species. Gene 643:124–132. https://doi.org/10.1016/j.gene.2017.12.00
Acknowledgements
We thank the French NGO Ailerons for providing fin clips of adult Prionacea glauca collected in the Mediterranean Sea, B. Holmes (University of Queensland) for providing us with Galeocerdo cuvier fin clips collected on the Great Barrier reef off the Australian coast, M. Meekan (Australian Institute of Marine Science) for providing Rhincodon typus samples collected on the Ningaloo reef off the west coast of Australia, and J. Ovenden (Southern Fisheries Center, Australia) for providing Carcharhinus sorrah samples collected on the Great Barrier reef. The Sphyrna lewini samples were collected by J. B. Galves in Madagascar near Nosy Be Island.
Funding
Funding was provided by École Pratique des Hautes Etudes (EPHE).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Ethical approval
Sampling of blacktip reef sharks was conducted by the Center for Island Research and Environmental Observatory (CRIOBE) under the permit N°9524 issued by the Ministère de la Promotion des Langues, de la Culture, de la Communication et de l’Environnement of the French Polynesian government in October 2015.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Eustache, K.B., Boissin, É., Tardy, C. et al. Characterization of 35 new microsatellite markers for the blacktip reef shark (Carcharhinus melanopterus) and cross-species amplification in eight other shark species. Mol Biol Rep 50, 3205–3215 (2023). https://doi.org/10.1007/s11033-022-08209-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08209-z