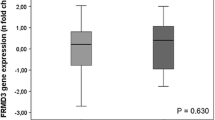
Abstract
Background
Long non-coding RNAs (lncRNAs) are key regulators of gene expression. Some studies have reported the association of polymorphisms in lncRNA genes with diabetes mellitus (DM) and its chronic complications, including diabetic kidney disease (DKD); however, the results are still inconclusive. Thus, we investigated the association of the rs3200401/MALAT1, rs1894720/MIAT, rs3931283/PVT1, rs11993333/PVT1, rs5749201/TUG1, and rs7158663/MEG3 polymorphisms with DKD in patients with type 2 DM (T2DM).
Methods and results
This study comprised 902 patients with T2DM and DKD (cases) and 394 patients with T2DM without DKD (controls). The six polymorphisms of interest were genotyped by real-time PCR using TaqMan probes. Frequency of the rs3931283/PVT1 G/G genotype was 36.2% in cases and 31.9% in controls (P = 0.331). After adjustment for gender, glycated hemoglobin, HDL cholesterol, ethnicity, hypertension, and diabetic retinopathy, the G/G genotype was associated with risk for DKD (OR = 1.625, 95% CI 1.020–2.588; P = 0.041). The rs3931283/PVT1 G/G genotype was also associated with higher urinary albumin excretion levels compared to A allele carriers (P = 0.017). No difference was found in rs7158663/MEG3 genotype frequencies between T2DM controls and DKD patients (OR = 1.087, 95% CI 0.686–1.724; P = 0.722). However, the rs7158663/MEG3 G/G genotype was associated with protection against severe DKD (OR = 0.694, 95% CI 0.488–0.989; P = 0.043, for patients with severe DKD vs. T2DM controls). The rs7158663/MEG3 G/G genotype was also associated with lower creatinine levels (P = 0.007) and higher estimated glomerular filtration rate (P = 0.010) compared to A allele carriers. No association was found between the rs11993333/PVT1, rs3200401/MALAT1, rs1894720/MIAT, and rs5749201/TUG1 polymorphisms and DKD or its laboratory markers.
Conclusion
The rs3931283/PVT1 G/G and rs7158663/MEG3 G/G are associated with DKD and markers of renal function in T2DM patients from a Brazilian population.

Similar content being viewed by others
References
Alicic RZ, Rooney MT, Tuttle KR (2017) Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clin J Am Soc Nephrol 12(12):2032 – 45.10.2215/CJN.11491116
Doshi SM, Friedman AN (2017) Diagnosis and Management of Type 2 Diabetic Kidney Disease. Clin J Am Soc Nephrol 12(8):1366 – 73.10.2215/CJN.11111016
Gu HF (2019) Genetic and Epigenetic Studies in Diabetic Kidney Disease. Front Genet 10:507.10.3389/fgene.2019.00507
Chen Y, He Y, Zhou H (2020) The potential role of lncRNAs in diabetes and diabetic microvascular complications. Endocr J 67(7):659 – 68.10.1507/endocrj.EJ19-0574
Jarroux J, Morillon A, Pinskaya M (2017) History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol 1008:1-46.10.1007/978-981-10-5203-3_1
Dieter C, Lemos NE, Correa NRF, Assmann TS, Crispim D (2021) The Impact of lncRNAs in Diabetes Mellitus: A Systematic Review and In Silico Analyses. Front Endocrinol (Lausanne) 12:602597.10.3389/fendo.2021.602597
Kato M, Natarajan R (2019) Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol 15(6):327 – 45.10.1038/s41581-019-0135-6
Hu M, Wang R, Li X, Fan M, Lin J, Zhen J et al (2017) LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with beta-catenin. J Cell Mol Med 21(11):2732 – 47.10.1111/jcmm.13189
Liu DW, Zhang JH, Liu FX, Wang XT, Pan SK, Jiang DK et al (2019) Silencing of long noncoding RNA PVT1 inhibits podocyte damage and apoptosis in diabetic nephropathy by upregulating FOXA1. Exp Mol Med 51(8):1-15.10.1038/s12276-019-0259-6
Yu D, Yang X, Zhu Y, Xu F, Zhang H, Qiu Z (2021) Knockdown of plasmacytoma variant translocation 1 (PVT1) inhibits high glucose-induced proliferation and renal fibrosis in HRMCs by regulating miR-23b-3p/early growth response factor 1 (EGR1). Endocr J 68(5):519 – 29.10.1507/endocrj.EJ20-0642
Zhong W, Zeng J, Xue J, Du A, Xu Y (2020) Knockdown of lncRNA PVT1 alleviates high glucose-induced proliferation and fibrosis in human mesangial cells by miR-23b-3p/WT1 axis. Diabetol Metab Syndr 12:33.10.1186/s13098-020-00539-x
Hanson RL, Craig DW, Millis MP, Yeatts KA, Kobes S, Pearson JV et al (2007) Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 56(4):975 – 83.10.2337/db06-1072
Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL et al (2016) Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest 126(11):4205 – 18.10.1172/JCI87927
Deng Q, Wen R, Liu S, Chen X, Song S, Li X et al (2020) Increased long noncoding RNA maternally expressed gene 3 contributes to podocyte injury induced by high glucose through regulation of mitochondrial fission. Cell Death Dis 11(9):814.10.1038/s41419-020-03022-7
Ji TT, Qi YH, Li XY, Tang B, Wang YK, Zheng PX et al (2020) Loss of lncRNA MIAT ameliorates proliferation and fibrosis of diabetic nephropathy through reducing E2F3 expression. J Cell Mol Med 24(22):13314 – 23.10.1111/jcmm.15949
von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP et al (2014) The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: guidelines for reporting observational studies. Int J Surg 12(12):1495 – 9.10.1016/j.ijsu.2014.07.013
Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E et al (2009) STrengthening the REporting of Genetic Association Studies (STREGA)--an extension of the STROBE statement. Genet Epidemiol 33(7):581 – 98.10.1002/gepi.20410
Canani LH, Capp C, Ng DP, Choo SG, Maia AL, Nabinger GB et al (2005) The fatty acid-binding protein-2 A54T polymorphism is associated with renal disease in patients with type 2 diabetes. Diabetes 54(11):3326 – 30.10.2337/diabetes.54.11.3326
American Diabetes A (2019) 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2019. Diabetes Care 42(Suppl 1):S13-S28.10.2337/dc19-S002
Andrassy KM (2013) Comments on ‘KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease’. Kidney international 84(3):622 – 3.10.1038/ki.2013.243
Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3, Feldman HI et al (2009) A new equation to estimate glomerular filtration rate. Ann Intern Med 150(9):604–612
de Souza BM, Michels M, Sortica DA, Boucas AP, Rheinheimer J, Buffon MP et al (2015) Polymorphisms of the UCP2 Gene Are Associated with Glomerular Filtration Rate in Type 2 Diabetic Patients and with Decreased UCP2 Gene Expression in Human Kidney. PLoS One 10(7):e0132938.10.1371/journal.pone.0132938
Camargo JL, Zelmanovitz T, Paggi A, Friedman R, Gross JL (1998) Accuracy of conversion formulae for estimation of glycohaemoglobin. Scand J Clin Lab Invest 58(6):521 – 8.10.1080/00365519850186337
Zelmanovitz T, Gross JL, Oliveira JR, Paggi A, Tatsch M, Azevedo MJ (1997) The receiver operating characteristics curve in the evaluation of a random urine specimen as a screening test for diabetic nephropathy. Diabetes Care 20(4):516 – 9.10.2337/diacare.20.4.516
Wilkinson CP, Ferris FL 3, Klein RE, Lee PP, Agardh CD, Davis M et al (2003) Proposed international clinical diabetic retinopathy and diabetic macular edema disease severity scales. Ophthalmology 110(9):1677 – 82.10.1016/S0161-6420(03)00475-5
Millis MP, Bowen D, Kingsley C, Watanabe RM, Wolford JK (2007) Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes 56(12):3027 – 32.10.2337/db07-0675
Ghaedi H, Zare A, Omrani MD, Doustimotlagh AH, Meshkani R, Alipoor S et al (2018) Genetic variants in long noncoding RNA H19 and MEG3 confer risk of type 2 diabetes in an Iranian population. Gene 675:265 – 71.10.1016/j.gene.2018.07.002
Rao SQ, Hu HL, Ye N, Shen Y, Xu Q (2015) Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr Res 166(1–3):125 – 30.10.1016/j.schres.2015.04.032
Duan J, Shen T, Dong H, Han S, Li G (2021) Association of the Expression Levels of Long-Chain Noncoding RNA TUG1 and Its Gene Polymorphisms with Knee Osteoarthritis. Genet Test Mol Biomarkers 25(2):102 – 10.10.1089/gtmb.2020.0208
Li Y, Zhang W, Ke H, Wang Y, Duan C, Zhu Q et al (2022) Rs1894720 polymorphism is associated with the risk of age-related cataract by regulating the proliferation of epithelial cells in the lens via the signalling pathway of MIAT/miR-26b/BCL2L2. Arch Med Sci 18(1):223 – 36.10.5114/aoms.2020.91533
Lahiri DK, Nurnberger JI (1991) A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res 19(19):5444
Zintzaras E, Lau J (2008) Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 61(7):634 – 45.10.1016/j.jclinepi.2007.12.011
Cao L, Qin P, Zhang J, Qiao H, Shi P, Huo H (2020) LncRNA PVT1 Suppresses the Progression of Renal Fibrosis via Inactivation of TGF-beta Signaling Pathway. Drug Des Devel Ther 14:3547 – 57.10.2147/DDDT.S245244
da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC (2008) Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet 24(6):306 – 16.10.1016/j.tig.2008.03.011
Zhang D, Qin H, Leng Y, Li X, Zhang L, Bai D et al (2018) LncRNA MEG3 overexpression inhibits the development of diabetic retinopathy by regulating TGF-beta1 and VEGF. Exp Ther Med 16(3):2337 – 42.10.3892/etm.2018.6451
Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG (2010) The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet 42(1):68-71.10.1038/ng.493
Zha F, Qu X, Tang B, Li J, Wang Y, Zheng P et al (2019) Long non-coding RNA MEG3 promotes fibrosis and inflammatory response in diabetic nephropathy via miR-181a/Egr-1/TLR4 axis. Aging (Albany NY) 11(11):3716 – 30.10.18632/aging.102011
Chang WW, Zhang L, Yao XM, Chen Y, Zhu LJ, Fang ZM et al (2020) Upregulation of long non-coding RNA MEG3 in type 2 diabetes mellitus complicated with vascular disease: a case-control study. Mol Cell Biochem 473(1–2):93 – 9.10.1007/s11010-020-03810-x
Su Y, Wu H, Pavlosky A, Zou LL, Deng X, Zhang ZX et al (2016) Regulatory non-coding RNA: new instruments in the orchestration of cell death. Cell Death Dis 7(8):e2333.10.1038/cddis.2016.210
Tello-Flores VA, Valladares-Salgado A, Ramirez-Vargas MA, Cruz M, Del-Moral-Hernandez O, Cahua-Pablo JA et al (2020) Altered levels of MALAT1 and H19 derived from serum or serum exosomes associated with type-2 diabetes. Noncoding RNA Res 5(2):71 – 6.10.1016/j.ncrna.2020.03.001
Li X, Zeng L, Cao C, Lu C, Lian W, Han J et al (2017) Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res 350(2):327 – 35.10.1016/j.yexcr.2016.12.006
Mohammad HMF, Abdelghany AA, Al Ageeli E, Kattan SW, Hassan R, Toraih EA et al (2021) Long Non-Coding RNAs Gene Variants as Molecular Markers for Diabetic Retinopathy Risk and Response to Anti-VEGF Therapy. Pharmacogenomics and personalized medicine 14:997-1014.10.2147/PGPM.S322463
Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang X et al (2018) Long non-coding RNA MIAT in development and disease: a new player in an old game. J Biomed Sci 25(1):23.10.1186/s12929-018-0427-3
Zhang M, Zhao S, Xu C, Shen Y, Huang J, Shen S et al (2020) Ablation of lncRNA MIAT mitigates high glucose-stimulated inflammation and apoptosis of podocyte via miR-130a-3p/TLR4 signaling axis. Biochem Biophys Res Commun 533(3):429 – 36.10.1016/j.bbrc.2020.09.034
Li Y, Xu K, Xu K, Chen S, Cao Y, Zhan H (2019) Roles of Identified Long Noncoding RNA in Diabetic Nephropathy. J Diabetes Res 2019:5383010.10.1155/2019/5383010
Zhou L, Xu DY, Sha WG, Shen L, Lu GY, Yin X (2015) Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem Biophys Res Commun 468(4):726 – 32.10.1016/j.bbrc.2015.11.023
Guo C, Qi Y, Qu J, Gai L, Shi Y, Yuan C (2020) Pathophysiological Functions of the lncRNA TUG1. Curr Pharm Des 26(6):688-700.10.2174/1381612826666191227154009
Zang XJ, Li L, Du X, Yang B, Mei CL (2019) LncRNA TUG1 inhibits the proliferation and fibrosis of mesangial cells in diabetic nephropathy via inhibiting the PI3K/AKT pathway. Eur Rev Med Pharmacol Sci 23(17):7519 – 25.10.26355/eurrev_201909_18867
Acknowledgements
This study was partially supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundo de Incentivo à Pesquisa e Eventos (FIPE) at Hospital de Clínicas de Porto Alegre (grant number: 2020 − 0656), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) (Edital FAPERGS/CNPq PRONEX 12/2014: 16-2551-0000476-5) ( FAPERGS 05/2019 – Programa Pesquisador Gaúcho PqG), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Graduate Program in Medical Sciences: Endocrinology – Universidade Federal do Rio Grande do Sul. D.C., and L.H.C are recipients of scholarship from CNPq; C.D. and F.M.P are recipient of scholarships from CAPES; and E.G. is a recipient of a scholarship from FAPERGS.
Author information
Authors and Affiliations
Contributions
Cristine Dieter and Daisy Crispim contributed to the study conception and design. Material preparation and data collection were performed by Cristine Dieter, Natália Emerim Lemos, Felipe Mateus Pellenz, Eliandra Girardi and Denise Taurino Ramos. Statistical Analysis was performed by Cristine Dieter, Natália Emerim Lemos, Taís Silveira Assmann and Daisy Crispim. Data Interpretation was performed by Cristine Dieter, Natália Emerim Lemos, Luís Henrique Canani, Taís Silveira Assmann, Daisy Crispim. The first draft of the manuscript was written by Cristine Dieter and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee in Research from Hospital de Clínicas de Porto Alegre (number 2020 − 0656). All subjects provided assent and written informed consent prior the inclusion in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dieter, C., Lemos, N.E., Girardi, E. et al. The rs3931283/PVT1 and rs7158663/MEG3 polymorphisms are associated with diabetic kidney disease and markers of renal function in patients with type 2 diabetes mellitus. Mol Biol Rep 50, 2159–2169 (2023). https://doi.org/10.1007/s11033-022-08122-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08122-5