Abstract
Background
Butter catfish (Ompok bimaculatus) is a preferred species in South East Asia, with huge aquaculture potential. However, there is limited information about genetic stock composition due to insufficient markers. The goal of this study was to develop de novo microsatellite markers.
Methods and results
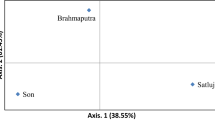
For sequencing, genomic SMRT bell libraries (1.5 Kbp size) were prepared for O. bimaculatus. A total of 114 SSR containing sequences were used for primer designing. Polymorphic loci were validated by genotyping 83 individuals from four distant riverine populations, viz., Brahmaputra, Bichiya, Gomti and Kaveri. A total of 30 microsatellite loci were polymorphic, of which five were found to be associated with functional genes and eight (four positive and four negative) loci were found to be under selection pressure. A total of 115 alleles were detected in all loci and PIC ranged from 0.539 to 0.927 and pair-wise FST values from 0.1267 to 0.26002 (p < 0.001), with an overall FST value of 0.17047, indicating the presence of population sub-structure. Cross-species transferability of 29 loci (96.67%) was successful in congener species, Ompok pabda.
Conclusion
The novel SSR markers developed in this study would facilitate stock characterization of natural populations, to be used in future selection breeding programs and planning conservation strategies in these species. Identified non-neutral markers will give insights into the effect of local adaptation on genetic differentiation in the natural population of this species.


Similar content being viewed by others
References
FAO (2021) Fishery and aquaculture statistics. Global capture production 1950–2019 (FishStatJ). FAO Fisheries Division (online)
Sarkar UK, Agnihotri P, Kumar R, Awasthi A, Pandey BK, Mishra A (2017) Dynamics of inter-population reproductive pattern in butter catfish, Ompok bimaculatus (Bloch, 1794) from different rivers in India. 17:1061–1071Turk J Fish Aquat Sci5
Molur S, Walker S (1998) Report of the Workshop “Conservation Assessment and Management Plan for Freshwater Fishes of India”, Zoo Outreach Organisation,Conservation Breeding Specialist Group, India, Coimbatore, India.156p
Ng HH, Tenzin K, Pal M(2010) Ompok bimaculatus The IUCN Red List of Threatened Species. 2010 RLTS.T166616A6248140.en
Liu ZJ, Cordes JF (2004) DNA marker technologies and their applications in aquaculture genetics. Aquaculture 242:735–736
Seth JK, Mohapatra A, Mohanty SR, Roy S (2021) Microsatellite Markers for Fish Conservation. In: Pandey PK, Parhi J (eds) Advances in Fisheries Biotechnology. Springer, Singapore, pp 175–181
Sigang F, Hao H, Yong L, Pengfei W, Chao Z, Lulu Y, Xiuting Q, Qiu L (2021) Genome-wide identification of microsatellite and development of polymorphic SSR markers for spotted sea bass (Lateolabrax maculatus). 20:100677Aquac Rep
Kirk H, Freeland JR (2011) Applications and implications of neutral versus non-neutral markers in molecular ecology. Int J Mol Sci 12(6):3966–3988
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877
Beier S, Thiel T, Münch T, Scholz U, Mascher M (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33(16):2583–2585
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. 40:e115–e115Nucleic Acids Res15
Pongsomboona S, Whanb V, Mooreb SS, Tassanakajona A (2000) Characterization of tri- and tetranucleotide microsatellites in the black tiger prawn, Penaeus monodon. Science Asia26:1–8
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16(5):1099–1106
Rousset F (2008) genepop’007: A complete re-implementation of the GENEPOP software for software for teaching and research. Mol Ecol Resour 8:103–106
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:117693430500100003
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Foll M, O Gaggiotti (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Stanke M, Morgenstern B (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33(2):W465-W467
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35(2):W182-W185
Huang K, Ritland K, Dunn DW, Qi X, Guo S, Li B (2016) Estimating relatedness in the presence of null alleles. Genetics 202(1):247–260
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43(1):223–225
Restrepo-Escobar N, Márquez EJ (2020) Microsatellite loci development for three catfish species from northwestern South America. Neotrop Ichthyol 18(1)
Acharya AP, Pavan-Kumar A, Gireesh-Babu P, Joshi CG, Chaudhari A, Krishna G (2019) Population genetics of Indian giant river-catfish, Sperataseenghala (Sykes, 1839) using microsatellite markers. Aquat Living Resour 32:4
Santos MDC, Ruffino ML, Farias IP (2007) High levels of genetic variability and panmixia of the tambaqui Colossoma macropomum (Cuvier, 1816) in the main channel of the Amazon River. J Fish Biol 71:33–44
Malakar AK, Lakra, WS, Goswami M, Mishra RM (2013) Genetic differentiation of Ompok bimaculatus (Teleostei: Siluridae) population based on mtDNA cytochrome b gene. Mitochondrial DNA 24:145–150
DeWoody JA, Avise JC (2000) Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J Fish Biol 56(3):461–473
Ferguson M(1994) The role of molecular genetic markers in the management of cultured fishes.Fish Biol Fish4:351–373
Torrente Y, Bella P, Tripodi L, Villa C, Farini A (2020) Role of insulin-like growth factor receptor 2 across muscle homeostasis: Implications for treating muscular dystrophy. Cells 9(2):441
Garcia de la, Serrana D, Macqueen DJ (2018) Insulin-like growth factor-binding proteins of teleost fishes. Front Endocrinol 9:80
Pellissier T, Al Nafea, H, Good, SV (2018) Divergence of insulin superfamily ligands, receptors and Igf binding proteins in marine versus freshwater stickleback: evidence of selection in known and novel genes. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics 25:53–61
Sen A, Madhivanan K, Mukherjee D, Aguilar RC(2012) The epsin protein family: coordinators of endocytosis and signaling.Biomol Concepts(2):117–126
Mettlen M, Chen PH, Srinivasan S, Danuser G, Schmid SL (2018) Regulation of clathrin-mediated endocytosis. Annu Rev Biochem 87:871–896
Wright S(1965) The interpretation of population structure by F-statistics with special regard to systems of mating.Evolution395–420
Koenig D, Hagmann J, Li R, Bemm F, Slotte T, Neuffer B, Wright SI, Weigel D (2019) Long-term balancing selection drives evolution of immunity genes in. Capsella eLife 8:e43606
Brandt DY, César J, Goudet J, Meyer D (2018) The effect of balancing selection on population differentiation: a Study with HLA Genes. Genes Genomes Genetics G3(8):2805–2815
Sahoo L, Barat A, Sahoo SK, Sahoo B, Das G, Das P, Sundaray JK, Swain SK (2020) Genetic diversity and population structure of endangered Indian catfish, Clarias magur as revealed by mtDNA D-loop marker. Turk J Fish & Aquat Sci21(1):09–18
Theodorou K, Couvet D (2015) The efficiency of close inbreeding to reduce genetic adaptation to captivity. Heredity 114(1):38–47
Brauer CJ, Hammer MP, Beheregaray LB (2016) Riverscape genomics of a threatened fish across a hydroclimatically heterogeneous river basin. Mol Ecol 25(20):5093–5113
Ward RD, Woodwark M, Skibinski DOF (1994) A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. 44:213–232J Fish Biol2
Agostinho AA, Pelicice FM, Gomes LC (2008) Dams and the fish fauna of the Neotropical region: impacts and management related to diversity and fisheries. 68:1119–1132Braz J Biol
Hatanaka T, Galetti PM Jr (2003) RAPD markers indicate the occurrence of structured populations in a migratory freshwater fish species. Genet Mol Biol 26:19–25
Wachirachaikarn A, Sutthakiet O, Senanan W, Na-Nakorn U (2020) Development of the new microsatellite multiplex PCR panel and genetic variation of farmed snakeskin gourami, Trichopodus pectoralis. 28:751–765Aquac Int2
Chowdhury LM, Kathirvelpandian A, Divya PR, Basheer VS, Mohitha C, Pavan-Kumar A, Krishna G (2021) Genetic characterization of Kiddi shrimp, Parapenaeopsis stylifera (H. Milne Edwards, 1837) along the Indian coast using microsatellite markers. 244:106128Fish Res
Alam J, Andriyono S, Hossain A, Eunus ATM, Kim HW(2019) The complete mitochondrial genome of a Pabdah catfish, Ompok pabda (Hamilton, 1822). Mitochondrial DNA Part B 4(1):507–508
Mandal S, Jena JK, Singh RK, Mohindra V, Lakra WS, Deshmukhe G Pathak A, Lal KK (2016) De novo development and characterization of polymorphic microsatellite markers in a schilbid catfish, Silonia silondia (Hamilton, 1822) and their validation for population genetic studies. Mol Biol Rep 43(2):91–98
Ali MF, Salam MA, Sarder MRI, Rahman MM, Mollah MFA (2021) Genetic Diversity and Population Structure of Endangered Catfish Rita rita (Hamilton, 1822) Revealed by Heterologous DNA Microsatellite Markers. Asian Fish Sci 34:181–194
Acknowledgements
The authors are thankful to The Director, National Bureau of Fish Genetic Resources (NBFGR), Lucknow for facilitating the work. The present work is the part of ICAR-NBFGR Project FISHNBFGRSIL202000900224, funded by Indian Council of Agricultural Research, New Delhi. Thanks are due to Dr. Ratnesh Tripathi for his help in generating long read sequences. Mr. Kantharajan is acknowledged for his role in preparing location map.
Funding
This investigation was supported by ICAR-National Bureau of Fish Genetic Resources, Lucknow, India, under project No. FISHNBFGRSIL202000900224.
Author information
Authors and Affiliations
Contributions
Kuldeep K. Lal conceived the idea and designed the experiments. Investigation, data collection and analysis were performed by Labrechai Mog Chowdhury, Shradha Chaturvedi, Sangeeta Mandal, Rajesh Kumar, Rajeev K. Singh and Vindhya Mohindra. The first draft of the manuscript was written by Sangeeta Mandal, Shradha Chaturvedi, Labrechai Mog Chowdhury, Vindhya Mohindra and all authors commented on previous versions of the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
The experiments were approved by Institute Animal Ethics Committee (IAEC), ICAR-NBFGR, Lucknow vide No. G/IAEC/2020/022 dated 09-02-2021.
Consent to participate and publish
All authors reviewed and approved the final version for publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chowdhury, L.M., Chaturvedi, S., Mandal, S. et al. Development of novel microsatellite markers for population differentiation and detection of natural selection in wild populations of butter catfish, Ompok bimaculatus (Bloch, 1794). Mol Biol Rep 50, 2435–2444 (2023). https://doi.org/10.1007/s11033-022-08105-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08105-6