Abstract
Background
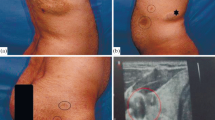
Neurofibromatosis type 1 (NF1) is an autosomal dominant with haploinsufficient, and multisystemic disorder including patches of skin Café-au-lait spots, Lisch nodules in the iris, and tumors in the peripheral nervous systems or fibromatous skin.
Methods
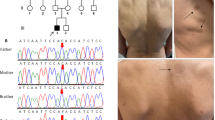
Blood samples were collected and DNA was extracted from a large Chinese pedigree suffering from NF1 disease with three spontaneous abortions or death for proband. Analysis for whole exome sequencing (WES), Sanger sequencing, and co-segregation was carried out. Prenatal gene diagnosis was also carried out in amniotic fluid DNA. The expression of NF1 was conducted by bioinformatics.
Results
A large Chinese pedigree with NF1 was recruited and a novel, heterozygous, variant (c.4272delA: p.I1426Ffs*2) for the NF1 gene in the proband was identified. This variant of NF1 produced a truncated protein that losses half of NF1 protein at the C-terminus including the CRAL-TRIO lipid-binding domain, NLS, and a small portion of Ras-GAP domain, thus leading to pathogenicity (ACMG criteria: PVS1 + PM2). NF1 expressions in different human tissues showed low tissue specificity, which may affect multiple organs presenting different phenotypes. Moreover, prenatal gene diagnosis for NF1 showed both alleles as wild types in the fetus of the proband.
Conclusion
We thus successfully identified a novel, pathogenic, heterozygous variant (c.4272delA:p.I1426Ffs*2) in the NF1 gene of NF1 disorder, expanding the NF1 mutation spectrum, that will help elucidate the molecular pathogenesis of NF1 disease and to contribute to the NF1 diagnosis, genetic counseling, clinical management in this large Chinese family.





Similar content being viewed by others
Data availability
All data used for the analyses of this study are available from the corresponding authors upon reasonable request.
References
Shannon KM, O’Connell P, Martin GA et al (1994) Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med 330:597–601
Sorensen SA, Mulvihill JJ, Nielsen A (1986) Long-term follow-up of von Recklinghausen neurofibromatosis. Survival and malignant neoplasms. New Engl J Med 314:1010–1015
Jin P, Yan K, Ye S et al (2021) Case Report: a synonymous mutation in NF1 located at the non-canonical splicing site leading to exon 45 skipping. Front Genet 12:772958
Bernier A, Larbrisseau A, Perreault S (2016) Cafe-au-lait macules and neurofibromatosis type 1: a review of the literature. Pediatr Neurol 60(24–9):e1
Alshahrani A, Abuoliat Z, Alshahrani AS et al (2022) Prevalence of associated endocrine diseases in patients with neurofibromatosis type 1. Avicenna J Med 12:16–20
Williams VC, Lucas J, Babcock MA et al (2009) Neurofibromatosis type 1 revisited. Pediatrics 123:124–133
Balcer LJ, Liu GT, Heller G et al (2001) Visual loss in children with neurofibromatosis type 1 and optic pathway gliomas: relation to tumor location by magnetic resonance imaging. Am J Ophthalmol 131:442–445
Ledbetter DH, Rich DC, O’Connell P et al (1989) Precise localization of NF1 to 17q11.2 by balanced translocation. Am J hum genet 44:20–24
Wallace MR, Andersen LB, Saulino AM et al (1991) A de novo Alu insertion results in neurofibromatosis type 1. Nature 353:864–866
Upadhyaya M, Osborn MJ, Maynard J et al (1997) Mutational and functional analysis of the neurofibromatosis type 1 (NF1) gene. Hum Genet 99:88–92
De Luca A, Bottillo I, Sarkozy A et al (2005) NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am J Hum Genet 77:1092–1101
Baralle D, Mattocks C, Kalidas K et al (2003) Different mutations in the NF1 gene are associated with Neurofibromatosis-Noonan syndrome (NFNS). Am J Med Genet A 119A:1–8
Trovo-Marqui AB, Tajara EH (2006) Neurofibromin: a general outlook. Clin Genet 70:1–13
Stewart DR, Brems H, Gomes AG et al (2014) Jaffe-Campanacci syndrome, revisited: detailed clinical and molecular analyses determine whether patients have neurofibromatosis type 1, coincidental manifestations, or a distinct disorder. Genet Med 16:448–459
Side LE, Emanuel PD, Taylor B et al (1998) Mutations of the NF1 gene in children with juvenile myelomonocytic leukemia without clinical evidence of neurofibromatosis, type 1. Blood 92:267–272
Ishida C, Gupta V. Genetics, Molecular Testing. StatPearls. Treasure Island (FL), 2021.
Sharifi S, Kalayci T, Palanduz S et al (2021) Clinical characteristics and mutation spectrum of neurofibromatosis type 1 in 27 Turkish families. Balkan Med J 38:365–373
Kehrer-Sawatzki H, Cooper DN (2022) Challenges in the diagnosis of neurofibromatosis type 1 (NF1) in young children facilitated by means of revised diagnostic criteria including genetic testing for pathogenic NF1 gene variants. Hum Genet 141:177–191
Fu J, Li L, Lu G (2002) Relationship between microdeletion on Y chromosome and patients with idiopathic azoospermia and severe oligozoospermia in the Chinese. Chin Med J 115:72–75
Cheng J, Fu J, Zhou Q et al (2019) A novel splicing mutation in the PRPH2 gene causes autosomal dominant retinitis pigmentosa in a Chinese pedigree. J Cell Mol Med 23:3776–3780
Wang F, Wang H, Tuan HF et al (2014) Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet 133:331–345
Zhang Q, Xu M, Verriotto JD et al (2016) Next-generation sequencing-based molecular diagnosis of 35 Hispanic retinitis pigmentosa probands. Sci Rep 6:32792
Imani S, Cheng J, Mobasher-Jannat A et al (2018) Identification of a novel RPGRIP1 mutation in an Iranian family with leber congenital amaurosis by exome sequencing. J Cell Mol Med 22:1733–1742
Fu Q, Xu M, Chen X et al (2017) CEP78 is mutated in a distinct type of Usher syndrome. J Med Genet 54:190–195
Koenekoop RK, Wang H, Majewski J et al (2012) Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat Genet 44:1035–1039
Uhlen M, Fagerberg L, Hallstrom BM et al (2015) Proteomics. Tissue-based map of the human proteome. Science 347:1260419
Fu J, Zhou B, Zhang L et al (2020) Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Mol Biol Rep 47:4383–4392
Cheng J, Peng J, Fu J et al (2020) Identification of a novel germline BRCA2 variant in a Chinese breast cancer family. J Cell Mol Med 24:1676–1683
Fu J, Cheng J, Liu X et al (2018) Evaluation genotypes of cancer cell lines HCC1954 and SiHa by short tandem repeat (STR) analysis and DNA sequencing. Mol Biol Rep 45:2689–2695
Zhu L, Cheng J, Zhou B et al (2017) Diagnosis for choroideremia in a large Chinese pedigree by nextgeneration sequencing (NGS) and noninvasive prenatal testing (NIPT). Mol Med Rep 15:1157–1164
Nykamp K, Anderson M, Powers M et al (2017) Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med 19:1105–1117
Dugoff L, Sujansky E (1996) Neurofibromatosis type 1 and pregnancy. Am J Med Genet 66:7–10
Remon-Ruiz P, Aliaga-Verdugo A, Guerrero-Vazquez R (2017) Pheochromocytoma in neurofibromatosis type 1 during pregnancy. Gynecol Endocrinol 33:93–95
Adams DR, Eng CM (2018) Next-generation sequencing to diagnose suspected genetic disorders. N Engl J Med 379:1353–1362
Acknowledgements
The authors thank the patients and family members for supporting our program.
Funding
ThE project was supported by the Technology Project Foundation of Luzhou City (Grant No. 2021-SYF-37), in part by the Research Foundation of the Science and Technology Department of Sichuan Province (Grant No. 2022NSFSC0737), and the National Natural Science Foundation of China (Grant Nos. 30371493, 31701087, and 81672887). We are thankful to Chiang Mai University, the Center for Research and Development of Natural Products for Health (Chiang Mai University).
Author information
Authors and Affiliations
Contributions
JF and SA were in charge of the idea, and project design. LY recruited samples. JiF, LY, XL, and JC performed DNA extraction, PCR amplification, and sequencing. JF and SA wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
None.
Ethical approval
The study has been approved by the Ethics Committee of Southwest Medical University in China. The informed consent form was obtained from the members of the family or guardian.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, L., Fu, J., Cheng, J. et al. Novel, heterozygous, pathogenic variant (c.4272delA: p.I1426Ffs*2) for the NF1 gene in a large Chinese family with neurofibromatosis type 1. Mol Biol Rep 50, 1117–1123 (2023). https://doi.org/10.1007/s11033-022-08096-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08096-4