Abstract
Background
The Cas9 nuclease is delivered in the form of either Cas9 protein or mRNA along with CRISPR guide RNA (gRNA: dual-crRNA:tracrRNA or chimeric single-guide RNA) or in a plasmid package encoding both Cas9 and the CRISPR gRNA.
Methods and results
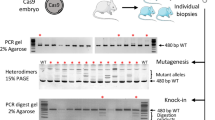
We directly compared the efficiency of producing rat blastocysts with homozygous mutations of the Foxn1 locus by pronuclear injection of Cas9 in the form of protein, mRNA, or plasmid DNA. For highly efficient production of rat blastocysts with homozygous Foxn1 mutations, pronuclear injection of Cas9 protein at 60 ng/µl was likely optimal. While blastocyst harvest in the mRNA groups was higher than those in the protein and plasmid DNA groups, genotype analysis showed that 63.6%, 8.7–20.0%, and 25.0% of the analyzed blastocysts were homozygous mutants in the protein, mRNA, and plasmid DNA groups, respectively. The high efficiency of producing homozygous mutant blastocysts in the 60 ng/µl protein group may be associated with primary genome editing being initiated before the first cleavage. In most cases, homozygous mutations at the target Foxn1 locus are triggered by deletion and repair via nonhomologous end joining or microhomology-mediated end joining. Deletion downstream of the Cas9 break site was more likely than deletion in the upstream direction.
Conclusions
The Cas9 nuclease in protein form, when coinjected with the CRISPR gRNA (ribonucleoprotein) into a rat zygote pronucleus, can access the target genome site and induce double-strand breaks promptly, resulting in the efficient production of homozygous mutants.

Similar content being viewed by others
References
Tong C, Huang G, Ashton C, Li P, Ying QL (2011) Generating gene knockout rats by homologous recombination in embryonic stem cells. Nat Protoc 6:827–844. https://doi.org/10.1038/nprot.2011.338
Behringer R, Gertsenstein M, Nagy KV, Nagy A (2014) Manipulating the mouse embryo: a laboratory manual, 4th edn. Cold Spring Harbor Laboratory Press, New York
Geurts AM, Cost GJ, Freyvert Y et al (2009) Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325:433. https://doi.org/10.1126/science.1172447
Tesson L, Usal C, Ménoret S et al (2011) Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 29:695–696. https://doi.org/10.1038/nbt.1940
Mashimo T, Kaneko T, Sakuma T et al (2013) Efficient gene targeting by TAL effector nucleases coinjected with exonucleases in zygotes. Sci Rep 3:1253. https://doi.org/10.1038/srep01253
Sung YH, Baek IJ, Kim DH et al (2013) Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 31:23–24. https://doi.org/10.1038/nbt.2477
Li W, Teng F, Li T, Zhou Q (2013) Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 31:684–686. https://doi.org/10.1038/nbt.2652
Shen B, Zhang J, Wu H et al (2013) Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23:720–723. https://doi.org/10.1038/cr.2013.46
Wang H, Yang H, Shivalila CS et al (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. https://doi.org/10.1016/j.cell.2013.04.025
Yeh CD, Richardson CD, Corn JE (2019) Advances in genome editing through control of DNA repair pathways. Nat Cell Biol 21:1468–1478. https://doi.org/10.1038/s41556-019-0425-z
Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109:E2579–E2586. https://doi.org/10.1073/pnas.1208507109
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Sung YH, Kim JM, Kim HT et al (2014) Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res 24:125–131. https://doi.org/10.1101/gr.163394.113
Aida T, Chiyo K, Usami T et al (2015) Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol 16:87. https://doi.org/10.1186/s13059-015-0653-x
Ménoret S, De Cian A, Tesson L et al (2015) Homology-directed repair in rodent zygotes using Cas9 and TALEN engineered proteins. Sci Rep 5:14410. https://doi.org/10.1038/srep14410
Nakagawa Y, Sakuma T, Nishimichi N et al (2016) Ultra-superovulation for the CRISPR-Cas9-mediated production of gene-knockout, single-amino-acid-substituted, and floxed mice. Biol Open 5:1142–1148. https://doi.org/10.1242/bio.019349
Yoshimi K, Kaneko T, Voigt B, Mashimo T (2014) Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat Commun 5:4240. https://doi.org/10.1038/ncomms5240
Li D, Qiu Z, Shao Y et al (2013) Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31:681–683. https://doi.org/10.1038/nbt.2661
Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M (2013) Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep 3:3355. https://doi.org/10.1038/srep03355
Goto T, Hara H, Sanbo M et al (2019) Generation of pluripotent stem cell-derived mouse kidneys in Sall1-targeted anephric rats. Nat Commun 10:451. https://doi.org/10.1038/s41467-019-08394-9
Yen ST, Zhang M, Deng JM et al (2014) Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol 393:3–9. https://doi.org/10.1016/j.ydbio.2014.06.017
Oliver D, Yuan S, McSwiggin H, Yan W (2015) Pervasive genotypic mosaicism in founder mice derived from genome editing through pronuclear injection. PLoS ONE 10:e0129457. https://doi.org/10.1371/journal.pone.0129457
Nowell CS, Bredenkamp N, Tetelin S et al (2011) Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PloS Genet 7:e1002348. https://doi.org/10.1371/journal.pgen.1002348
Goto T, Hara H, Nakauchi H, Hochi S, Hirabayashi M (2016) Hypomorphic phenotype of Foxn1 gene-modified rats by CRISPR/Cas9 system. Transgenic Res 25:533–544. https://doi.org/10.1007/s11248-016-9941-9
Sakurai T, Watanabe S, Kamiyoshi A, Sato M, Shindo T (2014) A single blastocyst assay optimized for detecting CRISPR/Cas9 system-induced indel mutations in mice. BMC Biotechnol 14:69. https://doi.org/10.1186/1472-6750-14-69
Bae S, Kweon J, Kim HS, Kim JS (2014) Microhomology-based choice of Cas9 nuclease target sites. Nat Methods 11:705–706. https://doi.org/10.1038/nmeth.3015
Abe T, Inoue KI, Furuta Y, Kiyonari H (2020) Pronuclear microinjection during S-phase increases the efficiency of CRISPR-Cas9-assisted knockin of large DNA donors in mouse zygotes. Cell Rep 31:107653. https://doi.org/10.1016/j.celrep.2020.107653
Kim S, Kim D, Cho SW, Kim J, Kim JS (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019. https://doi.org/10.1101/gr.171322.113
Ma Y, Chen W, Zhang X et al (2016) Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein. RNA Biol 13:605–612. https://doi.org/10.1080/15476286.2016.1185591
Santos F, Hendrich B, Reik W, Dean W (2002) Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 241:172–182. https://doi.org/10.1006/dbio.2001.0501
Taleei R, Nikjoo H (2013) Biochemical DSB-repair model for mammalian cells in G1 and early S phases of the cell cycle. Mutat Res 756:206–212. https://doi.org/10.1016/j.mrgentox.2013.06.004
Grajcarek J, Monlong J, Nishinaka-Arai Y et al (2019) Genome-wide microhomologies enable precise template-free editing of biologically relevant deletion mutations. Nat Commun 10:4856. https://doi.org/10.1038/s41467-019-12829-8
Vu TV, Doan DTH, Kim J et al (2021) CRISPR/Cas-based precision genome editing via microhomology-mediated end joining. Plant Biotechnol J 19:230–239. https://doi.org/10.1111/pbi.13490
Aida T, Nakade S, Sakuma T et al (2016) Gene cassette knock-in in mammalian cells and zygotes by enhanced MMEJ. BMC Genomics 17:979. https://doi.org/10.1186/s12864-016-3331-9
Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T (2016) ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun 7:10431. https://doi.org/10.1038/ncomms10431
Acknowledgements
The authors thank Reiko Terada, Fumika Yoshida, Mika Douki and Keiko Yamauchi (National Institute for Physiological Sciences) for their assistance with animal care and PCR sample preparations.
Funding
This work was supported by a grant from LEAP-AMED (JP18gm0010002) to M.H. and by the NINS program for Cross-Disciplinary Science study to T.G.
Author information
Authors and Affiliations
Contributions
TG: Conceptualization, Investigation, Data Curation, Writing—Original Draft, Funding Acquisition. KY: Investigation. SH: Data Curation, Validation, Visualization, Writing—Review & Editing. MH: Investigation, Data Curation, Writing—Review & Editing, Funding Acquisition, Supervision, Project Administration.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
All procedures for animal experimentation were reviewed and approved by the Animal Care and Use Committee of the National Institutes of Natural Sciences (Date: 2015.3.27/No. 15A102, Date: 2015.1.23/No. P09-070-A).
Consent to publish
All the authors have approved the manuscript being submitted for publication in the journal Molecular Biology Reports.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Goto, T., Yogo, K., Hochi, S. et al. Characterization of homozygous Foxn1 mutations induced in rat embryos by different delivery forms of Cas9 nuclease. Mol Biol Rep 50, 1231–1239 (2023). https://doi.org/10.1007/s11033-022-08054-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08054-0