Abstract
Background
The rice cultivars ASD 16 and ADT 43 are the most popular high-yielding Indica rice cultivars in southern India. Despite their popularity very little is known about their genetic basis due to lack of studies on the complete genome. In the current study, efforts were made to identify alleles and SNP markers that differentiate the two contrasting rice genotypes, ASD 16 and ADT 43 for grain shape and starch content.
Methods and results
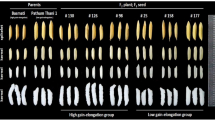
The complete genome of bold grain ASD 16 and slender grain ADT 43 were sequenced via Illumina’s paired-end sequencing and the reads obtained were mapped to the Oryza sativa Indica Group cultivar 93–11 reference genome. The grain size of rice is controlled by Quantitative Trait Loci (QTL) that has a robust effect on grain yield and quality. To gain insight into genes that controlling grain size and starch content, an in-silico analysis was performed by taking into account of 72 grain elongation and starch biosynthesis genes. The identified alleles were further validated in the whole genome sequencing data of 32 bold grain and 25 slender grain varieties that were retrieved from the 3 K rice genome project.
Conclusion
An “A to G” polymorphism leading to SER 74 PRO was identified at the CDS position 220 of the An-1 gene, encoding bHLH domain-containing protein that regulates awn formation and increase in grain length. The non-synonymous substitutions such as A545C variant leading PHE 182 CYS in ADP Glucose Pyrophosphorylase large subunit IV (AGPL4) and C3094G variant leading to VAL 1032 LEU in Starch synthase IIIb (OsSSIIIb) were also identified in the starch biosynthesis genes. These identified allelic variants may contribute to the crop improvement programs in rice.




Similar content being viewed by others
Data availability
ASD 16: The data from this study were submitted to NCBI under BioProject ID PRJNA666312 and BioSample ID SAMN16287940. The reads were submitted in compressed FastQ format at SRA database of NCBI with the accession number SRR12791393. ADT 43: Data were submitted to NCBI under BioProject ID PRJNA666316 and BioSample ID SAMN16288314. Quality reads were deposited in compressed FastQ format at SRA database of NCBI with the accession number SRR12777939.
Code availability
Not applicable.
References
Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends Genet 23:578–587
Singhabahu S, Wijesinghe C, Gunawardana D, Senarath-Yapa MD, Kannangara M, Edirisinghe R, Dissanayake VHW (2017) Whole genome sequencing and analysis of Godawee, a salt tolerant Indica rice variety. J Rice Res 5:177
Eckardt NA, Cominelli E, Galbiati M, Tonelli C (2009) The future of science: food and water for life. Plant Cell 21:368–372
FAO (2009) Declaration of the world summit on food security. UN, New York, pp 16–18
Sangeetha S, Kavitha P, Rajendran K (2013) Relative susceptibility of paddy varieties to Sitotroga cerealella Oliv. (Lepidoptera: Gelichiidae). Int J Res Phytochem Pharmacol 3:128–131
Saravanan S, Latha R, Arumugam Pillai M (2020) Studies on relative impact of rice varieties ASD 16 and TPS 5 on farmer’s adoption. Int J Curr Microbiol App Sci 9:1509–1513
Yang LV, Yueying W, Jahan N, Haitao H, Ping C, Lianguang S, Haiyan L, Guojun D, Jiang H, Zhenyu G, Qian Q, Yu Z, Longbiao G (2019) Genome-wide association analysis and allelic mining of grain shape-related traits in rice. Rice Sci 26:384–392
Huang H, Qian Q (2017) Progress in genetic research of rice grain shape and breeding achievements of long-grain shape and good quality japonica rice. Chin J Rice Sci 31:665–672
Huang R, Jiang L, Zheng J, Wang T, Wang H, Huang Y, Hong Z (2013) Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci 18:218–226
Sun Y, Yan L, Dong C, Wang P, Huang X, Deng X (2005) Genetic relationship among Wx gene, AC, GC and GT of rice. Acta Agron Sin 31:535–539
Bao JS (2014) Genes and QTLs for rice grain quality improvement. In: Yan W (ed) Rice—germplasm genetics and improvement. InTech-Open Science, London, pp 239–278
Cuevas RP, Fitzgerald MA (2012) Genetic diversity of rice grain quality. In: Caliskan M (ed) Genetic diversity in plants. InTech, London, pp 286–310
Dela Cruz N, Khush GS (2000) Rice grain quality evaluation procedures. In: Singh K, Khush GS (eds) Aromatic rices. Mohan Primlani for Oxford & IBH Publishing Co., New Delhi, pp 16–28
He P, Li SG, Qian Q, Ma YQ, Li JZ, Wang WM, Chen Y, Zhu LH (1999) Genetic analysis of rice grain quality. Theor Appl Genet 98:502–508
Bao JS, Zheng XW, Xia YW, He P, Shu QY, Lu X, Chen Y, Zhu LH (2000) QTL mapping for the paste viscosity in rice (Oryza sativa L.). Theor Appl Genet 100:280–284
Bao JS, Corke H, He P, Zhu LH (2003) Analysis of quantitative trait loci for starch properties of rice based on an RIL population. Act Bot Sin 45:986–994
Zhang C, Hu B, Zhu K, Zhang H, Leng Y, Tang S, Gu M, Liu Q (2013) QTL mapping for rice RVA properties using high-throughput re-sequenced chromosome segment substitution lines. Rice Sci. 20:407–414
Tran NA, Daygon VD, Resurreccion AP, Cuevas RP, Corpuz HM, Fitzgerald MA (2011) A single nucleotide polymorphism in the waxy gene explains a significant component of gel consistency. Theor Appl Genet 123:519–525
Zhang L, Ma B, Bian Z, Li X, Zhang C, Liu J, Li Q, Liu Q, He Z (2020) Grain size selection using novel functional markers targeting 14 genes in rice. Rice 13:63
Niu Y, Chen T, Wang C, Chen K, Shen C, Chen H, Zhu S, Wu Z, Zheng T, Zhang F, Xu J (2021) Identification and allele mining of new candidate genes underlying rice grain weight and grain shape by genome-wide association study. BMC Genomics 22:602
Ponce K, Zhang Y, Guo L, Leng Y, Ye G (2020) Genome-wide association study of grain size traits in indica rice multiparent advanced generation intercross (MAGIC) population. Front Plant Sci 11:395
Wu L, Cui Y, Xu Z, Xu Q (2020) Identification of multiple grain shape-related loci in rice using bulked segregant analysis with high-throughput sequencing. Front Plant Sci 11:303
Meng B, Wang T, Luo Y, Guo Y, Xu D, Liu C, Zou J, Li L, Diao Y, Gao Z, Hu Z, Zheng X (2022) Identification and allele combination analysis of rice grain shape-related genes by genome-wide association study. Int J Mol Sci 23(3):1065
Zheng C, Boer MP, van Eeuwijk FA (2015) Reconstruction of genome ancestry blocks in multiparental populations. Genetics 200:1073–1087
Chen H, Xie W, He H, Yu H, Chen W, Li J, Yu R, Yao Y, Zhang W, He Y, Tang X, Zhou F, Deng XW, Zhang Q (2014) A high-density SNP genotyping array for rice biology and molecular breeding. Mol Plant 7:541–553
Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC (2016) SIFT missense predictions for genomes. Nat Protoc 11:1–9
Wu HP, Wei FJ, Wu CC, Lo SF, Chen LJ, Fan MJ, Chen S, Wen IC, Yu SM, Ho TD, Lai MH, Hsing YC (2017) Large-scale phenomics analysis of a T-DNA tagged mutant population. Gigascience 6:1–7
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Kwon CT, Paek NC (2016) Gibberellic acid: a key phytohormone for spikelet fertility in rice grain production. Int J Mol Sci 17(5):794
Luo J, Liu H, Zhou T, Gu B, Huang X, Shangguan Y, Zhu J, Li Y, Zhao Y, Wang Y, Zhao Q, Wang A, Wang Z, Sang T, Han B (2013) An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25:3360–3376
Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K, Zhang X, Guo X, Wang J, Wang H, Wan J (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants 3:17043
Dian W, Jiang H, Wu P (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56:623–632
Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y (1999) The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol 121:399–410
Tuncel A, Kawaguchi J, Ihara Y, Matsusaka H, Nishi A, Nakamura T, Kuhara S, Hirakawa H, Nakamura Y, Cakir B, Nagamine A, Okita TW, Hwang SK, Satoh H (2014) The rice endosperm ADP-glucose pyrophosphorylase large subunit is essential for optimal catalysis and allosteric regulation of the heterotetrameric enzyme. Plant Cell Physiol 55:1169–1183
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Andrews S (2010) FastQC: A quality control tool for high throughput sequence data.
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Cingolani P, Platts A, le Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92
Chen E, Huang X, Tian Z, Wing RA, Han B (2019) The Genomics of Oryza species provides insights into rice domestication and heterosis. Annu Rev Plant Biol 70:639–665
Li X, Wei Y, Li J, Yang F, Chen Y, Guo S, Sha A (2020) Identification of QTL TGW12 responsible for grain weight in rice based on recombinant inbred line population crossed by wild rice (Oryza minuta) introgression line K1561 and indica rice G1025. BMC Genet 21:10
Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–549
Shi C, Ren Y, Liu L, Wang F, Zhang H, Tian P, Pan T, Wang Y, Jing R, Liu T, Wu F, Lin Q, Lei C, Zhang X, Zhu S, Guo X, Wang J, Zhao Z, Zhai H, Cheng Z, Wan J (2019) Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol 180:381–391
Wang S, Li S, Liu Q, Wu K, Zhang J, Wang Y, Chen X, Zhang Y, Gao C, Wang F, Huang H, Fu X (2015) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47:949–954
Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56:3229–3244
RGP (2014) The 3000 rice genomes project. Gigascience 3:7
Bolser DM, Staines DM, Perry E, Kersey PJ (2016) Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomic data. Methods Mol Biol 1533:1–31
Amid C, Alako BTF, Balavenkataraman Kadhirvelu V, Burdett T, Burgin J, Fan J, Harrison PW, Holt S, Hussein A, Ivanov E, Jayathilaka S, Kay S, Keane T, Leinonen R, Liu X, Martinez-Villacorta J, Milano A, Pakseresht A, Rahman N, Rajan J, Reddy K, Richards E, Smirnov D, Sokolov A, Vijayaraja S, Cochrane G (2019) The European nucleotide archive in 2019. Nucleic Acids Res 48:D70–D76
Bindusree G, Natarajan P, Kalva S, Madasamy P (2017) Whole genome sequencing of Oryza sativa L. cv. Seeragasamba identifies a new fragrance allele in rice. PLoS One 12:e0188920
Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD (2020) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49:D344–D354
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259
Yang Z, Rannala B (2012) Molecular phylogenetics: principles and practice. Nat Rev Genet 13:303–314
Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302
Leigh JW, Bryant D (2015) Popart: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Acknowledgements
This research was carried out in the Bioinformatics Laboratory, Dept. of Plant Molecular Biology and Bioinformatics, Centre for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University supported by the Biotechnology Information System (BTIS) program (No.BT/BI/04/015/89 Vol.III dated 13.03.2018 and BT/PR40235/BTIS/137/39/2022 dated 15.03.2022) of the Department of Biotechnology (DBT), Government of India, New Delhi.
Funding
The financial support of the Department of Biotechnology, Government of India through the program support for research and development in agricultural bioinformatics (BT/PR40235/BTIS/137/39/2022) is acknowledged.
Author information
Authors and Affiliations
Contributions
JR, MJ, RG and SM: Designed and directed the project. MJ, JR and MR: Performed genome sequencing. MLA, HDL, RS, and MJ: Performed data analysis. MJ, MR, MLA and LA: Drafted the manuscript and designed the figures. All authors discussed the result and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interest
The author(s) declare no competing interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mannu, J., Latha, A.M., Rajagopal, S. et al. Whole genome sequencing of ASD 16 and ADT 43 to identify predominant grain size and starch associated alleles in rice. Mol Biol Rep 49, 11743–11754 (2022). https://doi.org/10.1007/s11033-022-07935-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07935-8