Abstract
Background
Tetanops sintenisi is a pest that mainly damages the root of quinoa (Chenopodium quinoa) and it is first discovered in China in 2018.
Methods and Results
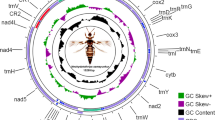
Here, the complete mitochondrial genome (mitogenome) of T. sintenisi was sequenced and compared with the mitogenomes of other Diptera species. The results revealed that the mitogenome of T. sintenisi is 15,763 bp in length (GenBank accession number: MT795181) and is comprised of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA genes, and a non-coding A + T-rich region (959 bp). The highly conserved gene arrangement of the mitogenome of T. sintenisi was identical to that of other Diptera insects. Twelve PCGs contained the typical insect start codon ATN, while cox1 had CGA as the start codon. The genes cox2, nad4, and nad1 contained an incomplete termination codon T; nad3, nad5, and cob contained the complete termination codon TAG; and the remaining seven PCGs contained the termination codon TAA. All tRNA genes were predicted to fold into the typical cloverleaf secondary structure. Phylogenetic analysis of 48 species based on the mitogenome sequence revealed that T. sintenisi clustered with the Tephritidae family, indicating that T. sintenisi and Tephritidae have a close phylogenetic relationship.
Conclusions
The phylogenetic relationship of T. sintenisi based on the mitogenome is consistent with the traditional morphological taxonomy, according to which T. sintenisi belongs to the family Otitidae, which is closely related to the family Muscidae.




Similar content being viewed by others
Data Availability
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/genbank under the accession no. MT795181. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA757578, SRR15725549, and SAMN20968555, respectively.
References
Zhang NX, Zhang YJ, Guo YU, Chen B (2013) Structure characteristics of the mitochondrial genomes of Diptera and design and application of universal primers for their sequencing. Acta Entomol Sin 56(4):398–407. https://doi.org/10.16380/j.kcxb.2013.04.013
Li H, Li J (2019) The complete mitochondrial genome of Helophilus virgatus (Diptera: Syrphidae: Eristalinae) with a phylogenetic analysis of Syrphidae. Mitochondrial DNA Part B 4(2):3106–3107. https://doi.org/10.1080/23802359.2019.1667890
Yan W, Shang Y, Ren L, Zhang X, Wang Y (2019) The complete mitochondrial genome of Hydrotaea dentipes (Diptera: Muscidae). Mitochondrial DNA Part B 4(1):2044–2045. https://doi.org/10.1080/23802359.2019.1618217
Zhang NX, Guo Y, Li TJ, He QY, Zhou Y, Si FL, Shuang R, Chen B, Yue BS (2015) The complete mitochondrial genome of Delia antiqua and its implications in Dipteran Phylogenetics. PLoS ONE 10(10):e0139736. https://doi.org/10.1371/journal.pone.0139736
Choudhary JS, Naaz N, Prabhakar CS, Rao MS, Das B (2015) The mitochondrial genome of the peach fruit fly, Bactrocera zonata (Saunders) (Diptera: Tephritidae): complete DNA sequence, genome organization, and phylogenetic analysis with other tephritids using next generation DNA sequencing. Gene 569(2):191–202. https://doi.org/10.1016/j.gene.2015.05.066
Wang T, Ren YL, Zhong Y, Yang MF (2020) Sequencing and analysis of the complete mitochondrial genome of Bactrocera cheni from China and its phylogenetic analysis. Mitochondrial DNA Part B 5(1):780–781. https://doi.org/10.1080/23802359.2020.1715882
Mitchell SE, Cockburn AF, Seawright JA (1993) The mitochondrial genome of Anopheles quadrimaculatus species A: complete nucleotide sequence and gene organization. Genome 36(6):1058–1073. https://doi.org/10.1139/g93-141
Spanos L, Koutroumbas G, Kotsyfakis M, Louis C (2010) The mitochondrial genome of the Mediterranean fruit fly, Ceratitis capitata. Insect Mol Biol 9(2):139–144. https://doi.org/10.1046/j.1365-2583.2000.00165.x
Lessinger AC, Junqueira A, Lemos TA, Kemper EL, Azeredo-Espin A (2010) The mitochondrial genome of the primary screwworm fly Cochliomyia hominivorax (Diptera: Calliphoridae). Insect Mol Biol 9(5):521–529. https://doi.org/10.1046/j.1365-2583.2000.00215.x
Stevens JR, West H, Wall R (2010) Mitochondrial genomes of the sheep blowfly, Lucilia sericata, and the secondary blowfly, Chrysomya megacephala. Med Vet Entomol 22(1):89–91. https://doi.org/10.1111/j.1365-2915.2008.00710.x
Junqueira A, Lessinger AC, Torres TT, Silva F, Espin A (2004) The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene 339(1):7–15. https://doi.org/10.1016/j.gene.2004.06.031
Beckenbach AT, Joy JB (2009) Evolution of the mitochondrial genomes of Gall midges (Diptera: Cecidomyiidae): rearrangement and severe truncation of tRNA genes. Genome Biol Evol 1(1):278–287. https://doi.org/10.1093/gbe/evp027
Oliveira MT, Barau JG, Junqueira A, Feijo PC, Lessinger AC (2008) Structure and evolution of the mitochondrial genomes of Haematobia irritans and Stomoxys calcitrans: the Muscidae (Diptera: Calyptratae) perspective. Mol Phylogenet Evol 48(3):850–857. https://doi.org/10.1016/j.ympev.2008.05.022
Clary DO, Wolstenholme DR (1985) The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol 22(3):252–271. https://doi.org/10.1007/BF02099755
Smit JT (2005) De prachtvlieg Tetanops sintenisi nieuw voor Nederland (Diptera: Ulidiidae). Nederlandse Faunistische Mededelingen 22:91–94
Xing K, Fei Z, Zhao X, Hui Y, Zhou J, Hong L (2018) First report of root maggot Tetanops sintenisi on quinoa. China Plant Protection 38(12):38–40
Mortelmans J, Bree ED, Hendrix J (2012) Four new additions to the Belgian fauna (Diptera: Conopidae, Tabanidae, Sciomyzidae, Ulidiidae). Bull van de Koninklijke Belgische vereniging voor entomologie 148:193–196
Stuke JH, Merz B (2005) Drei für Deutschland neu nachgewiesene acalyptrate Fliegen (Diptera: Lauxaniidae, Pallopteridae, Ulidiidae). Studia Dipterologica 12:242–254
Stuke JH (2006) Bemerkenswerte Zweiflügler aus Niedersachsen und Bremen (Insecta: Diptera) – 1. Teil Drosera 12:67–72
Stuke JH (2008) Die Tephritoidea (Diptera) Niedersachsens und Bremens. Abh. Naturwiss Vereins Bremen 46(2):329–356
John TS, Gerrit T (2014) Een bijzondere vangst van een bijzondere prachtvlieg (Diptera, Ulidiidae). Entomol Berichten 74(6):261–263
Coil D, Jospin G, Darling AE (2014) A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31(4):587–589. https://doi.org/10.1093/bioinformatics/btu661
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Kurtz S, Phillippy A, Delcher AL, Smoot M (2004) Versatile and open software for comparing large genomes. Genome Biol 5(2):R12. https://doi.org/10.1186/gb-2004-5-2-r12
Walker BJ, Abeel T, Shea T, Priest M, Earl AM (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9(11):e112963. https://doi.org/10.1371/journal.pone.0112963
Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69(2):313–319. https://doi.org/10.1016/j.ympev.2012.08.023
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinformatics 21(4):537–539. https://doi.org/10.1093/bioinformatics/bti054
Sudhir K, Glen S, Li M, Christina K, Koichiro T (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Narakusumo RP, Riedel A, Pons J (2020) Mitochondrial genomes of twelve species of hyperdiverse Trigonopterus weevils. PeerJ 8:e10017. https://doi.org/10.7717/peerj.10017
Lewis DL, Farr CL, Kaguni LS (2010) Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol Biol 4(4):263–278. https://doi.org/10.1111/j.1365-2583.1995.tb00032.x
Nardi F, Carapelli A, Dallai R, Frati F (2003) The mitochondrial genome of the olive fly Bactrocera oleae: two haplotypes from distant geographical locations. Insect Mol Biol 12(6):605–611. https://doi.org/10.1046/j.1365-2583.2003.00445.x
Brujn MHLD (1983) Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature 304(5923):234–241. https://doi.org/10.1038/304234a0
Kim I, Lee EM, Seol KY, Yun EY, Jin BR (2010) The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol Biol 15(2):217–225. https://doi.org/10.1111/j.1365-2583.2006.00630.x
Coates BS, Sumerford DV, Hellmich RL, Lewis LC (2005) Partial mitochondrial genome sequences of Ostrinia nubilalis and Ostrinia furnicalis. Int J Biol Sci 1(1):13–18. https://doi.org/10.7150/ijbs.1.13
Wang JP, Cao TW, Zhang Y, Fan RJ, Zhang M, Shi BM, Peng FC (2017) Sequencing and analysis of the complete mitochondrial genome of Limenitis helmanni (Lepidoptera: Nymphalidae). Acta Entomol Sin 60(8):950–961. https://doi.org/10.16380/j.kcxb.2017.08.012
Li T, Yang J, Li Y, Cui Y, Xie Q, Bu W, Hillis DM (2016) A mitochondrial genome of Rhyparochromidae (Hemiptera: Heteroptera) and a comparative analysis of related mitochondrial genomes. Sci Rep 6:35175. https://doi.org/10.1038/srep35175
Sun H, Zhao W, Lin R, Zhou Z, Huai W, Yao Y (2020) The conserved mitochondrial genome of the jewel beetle (Coleoptera: Buprestidae) and its phylogenetic implications for the suborder Polyphaga. Genomics 112(5):3713–3721. https://doi.org/10.1016/j.ygeno.2020.04.026
Oliveira M, Lessinger AC (2005) Evolutionary and structural analysis of the cytochrome c oxidase subunit I (COI) gene from, and (Diptera: Muscidae) mitochondrial DNA. Mitochondrial DNA Part A 16(2):156–160. https://doi.org/10.1080/10425170500039901
Yang F, Du YZ, Wang LP, Cao JM, Yu WW (2011) The complete mitochondrial genome of the leafminer Liriomyza sativae (Diptera: Agromyzidae): great difference in the A + T-rich region compared to Liriomyza trifolii. Gene 485(1):7–15. https://doi.org/10.1016/j.gene.2011.05.030
Jin SB, Kim I, Sohn HD, Jin BR (2004) The mitochondrial genome of the firefly, Pyrocoelia rufa: complete DNA sequence, genome organization, and phylogenetic analysis with other insects. Mol Phylogenet Evol 32(3):978–985. https://doi.org/10.1016/j.ympev.2004.03.009
Yue Z, Feng S, Zeng Y, Hong N, Liu L, Zhao Z, Fan J, Li Z (2018) The first complete mitochondrial genome of Bactrocera tsuneonis (Miyake) (Diptera: Tephritidae) by next-generation sequencing and its phylogenetic implications. Int J Biol Macromol 118(15):1229–1237. https://doi.org/10.1016/j.ijbiomac.2018.06.099
Su Y, Zhang Y, Feng S, He J, Zhao Z, Bai Z, Liu L, Zhang R, Li Z (2017) The mitochondrial genome of the wolfberry fruit fly, Neoceratitis asiatica (Becker) (Diptera: Tephritidae) and the phylogeny of Neoceratitis Hendel genus. Sci Rep 7(1):16612. https://doi.org/10.1038/s41598-017-16929-7
Ojala D, Montoya J, Attardi G (1981) Trna punctuation model of rna processing in human mitochondria. Nature 290(5806):470–474. https://doi.org/10.1038/290470a0
Zhang DX, Hewitt GM (1997) Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol 25(2):99–120. https://doi.org/10.1016/S0305-1978(96)00042-7
Brian MW, Michelle DT, Isaac SW, Norman BB, Jung-Wook K, Christine L, Matthew AB, Brian KC, Keith MB, Alysha MH, Benjamin MW, Kevin JP, Thomas P, Bradley JS, Jeffrey HS, Vladimir B, Jason C, Sujatha NK, Urs S-O, Gail EK, Christian F, David T, Andrew AG, Gregory TB, Markus WC, Rudolf F, David M KY (2011) Episodic radiations in the fly tree of life. PNAS 108(14):5690–5695. https://doi.org/10.1073/pnas.1012675108
Acknowledgements
We thank Wenna Wang for assistance in completing the experiments. This research was financially supported by the construction of modern agricultural (coarse cereals) industrial technology system in Shanxi Province (2022-03), the Foundation of Shanxi Academy of Agricultural Sciences (YCX2018D2T02) and the Province Key Research and Development Program of Shanxi (201903D221014).
Author information
Authors and Affiliations
Contributions
Conceptualization, K.X.; Formal analysis, K.X. and C.K.; Funding acquisition, F.Z.; Methodology, K.X and C.K.; Project administration, F.Z.; Writing-original draft, K.X. and C.K.; Writing-review and editing, X.J.Z and F.Z. All authors have read and agreed to the published version of the manuscript. Conflict of interest The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xing, K., Kang, C. & Zhao, F. The first complete mitochondrial genome sequences for Ulidiidae and phylogenetic analysis of Diptera. Mol Biol Rep 50, 2501–2510 (2023). https://doi.org/10.1007/s11033-022-07869-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07869-1