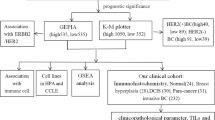
Abstract
Background
An emerging component of Unfolded Protein Response (UPR) pathway, cation transport regulator homolog 1 (CHAC1) has been conferred with the ability to degrade intracellular glutathione and induce apoptosis, however, many reports have suggested a role of CHAC1 in cancer progression. Our study aimed to investigate CHAC1 mRNA levels in large breast cancer datasets using online tools and both mRNA and protein levels in different breast cancer cell lines.
Methods and results
Analysis of clinical information from various online tools (UALCAN, GEPIA2, TIMER2, GENT2, UCSCXena, bcGenExMiner 4.8, Km Plotter, and Enrichr) was done to elucidate the CHAC1 mRNA expression in large breast cancer patient dataset and its correlation with disease progression. Later, in vitro techniques were employed to explore the mRNA and protein expression of CHAC1 in breast cancer cell lines. Evidence from bioinformatics analysis as well as in vitro studies indicated a high overall expression of CHAC1 in breast tumor samples and had a significant impact on the prognosis and survival of patients. Enhanced CHAC1 levels in the aggressive breast tumor subtypes such as Human Epidermal growth factor receptor 2 (HER2) and Triple Negative Breast Cancer (TNBC) were evident. Our findings hint toward the possible role of CHAC1 in facilitating the aggressiveness of breast cancer and the disease outcome.
Conclusion
In summary, CHAC1 is constantly up-regulated in breast cancer leading to a poor prognosis. CHAC1, therefore, could be a promising candidate in the analysis of breast cancer diagnosis and prognosis.





Similar content being viewed by others
Data availability
The datasets used in the present study are available in the public database and have been listed in the manuscript.
Abbreviations
- BC:
-
Breast cancer
- CHAC1:
-
Cation transport regulator homolog 1
- DMFS:
-
Distant metastasis free survival
- HER2:
-
Human epidermal growth factor receptor 2
- RFS:
-
Recurrence-free survival
- TNBC:
-
Triple-negative breast cancer
- UPR:
-
Unfolded Protein Response
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Urra H, Dufey E, Avril T, Chevet E, Hetz C (2016) Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer 2(5):252–262
Muz B, de la Puente P, Azab F, Azab AK (2015) The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 3:83
Mehta V, Chander H, Munshi A (2021) Complex roles of discoidin domain receptor tyrosine kinases in cancer. Clin Transl Oncol 23(8):1497–1510
McGrath EP, Logue SE, Mnich K, Deegan S, Jäger R, Gorman AM, Samali A (2018) The unfolded protein response in breast cancer. Cancers 10(10):344
Binet F, Sapieha P (2015) ER stress and angiogenesis. Cell Metab 22(4):560–575
Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y (2014) XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature 508(7494):103–107
Zhang K, Liu H, Song Z, Jiang Y, Kim H, Samavati L, Nguyen HM, Yang Z-Q (2020) The UPR transducer IRE1 promotes breast cancer malignancy by degrading tumor suppressor microRNAs. Iscience 23(9):101503
Logue SE, McGrath EP, Cleary P, Greene S, Mnich K, Almanza A, Chevet E, Dwyer RM, Oommen A, Legembre P (2018) Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat Commun 9(1):1–14
Zhao N, Cao J, Xu L, Tang Q, Dobrolecki LE, Lv X, Talukdar M, Lu Y, Wang X, Hu DZ (2018) Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J Clin Investig 128(4):1283–1299
Zhang X, Zhou Y, Mao F, Lin Y, Shen S, Sun Q (2020) lncRNA AFAP1-AS1 promotes triple negative breast cancer cell proliferation and invasion via targeting miR-145 to regulate MTH1 expression. Sci Rep 10(1):1–11
Feng Y-X, Jin DX, Sokol ES, Reinhardt F, Miller DH, Gupta PB (2017) Cancer-specific PERK signaling drives invasion and metastasis through CREB3L1. Nat Commun 8(1):1–10
Chi Z, Zhang J, Tokunaga A, Harraz MM, Byrne ST, Dolinko A, Xu J, Blackshaw S, Gaiano N, Dawson TM (2012) Botch promotes neurogenesis by antagonizing notch. Dev Cell 22(4):707–720
Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ (2009) CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol 182(1):466–476
Chen M-S, Wang S-F, Hsu C-Y, Yin P-H, Yeh T-S, Lee H-C, Tseng L-M (2017) CHAC1 degradation of glutathione enhances cystine-starvation-induced necroptosis and ferroptosis in human triple negative breast cancer cells via the GCN2-eIF2α-ATF4 pathway. Oncotarget 8(70):114588
Goebel G, Berger R, Strasak A, Egle D, Müller-Holzner E, Schmidt S, Rainer J, Presul E, Parson W, Lang S (2012) Elevated mRNA expression of CHAC1 splicing variants is associated with poor outcome for breast and ovarian cancer patients. Br J Cancer 106(1):189–198
Liu Y, Li M, Shi D, Zhu Y (2019) Higher expression of cation transport regulator-like protein 1 (CHAC1) predicts of poor outcomes in uveal melanoma (UM) patients. Int Ophthalmol 39(12):2825–2832
Ogawa T, Wada Y, Takemura K, Board PG, Uchida K, Kitagaki K, Tamura T, Suzuki T, Tokairin Y, Nakajima Y (2019) CHAC1 overexpression in human gastric parietal cells with Helicobacter pylori infection in the secretory canaliculi. Helicobacter 24(4):e12598
Wada Y, Takemura K, Tummala P, Uchida K, Kitagaki K, Furukawa A, Ishige Y, Ito T, Hara Y, Suzuki T (2018) Helicobacter pylori induces somatic mutations in TP 53 via overexpression of CHAC 1 in infected gastric epithelial cells. FEBS Open Bio 8(4):671–679
Parrales A, Iwakuma T (2015) Targeting oncogenic mutant p53 for cancer therapy. Front Oncol 5:288
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47(W1):W556–W560
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, Varambally S (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19(8):649–658
Park S-J, Yoon B-H, Kim S-K, Kim S-Y (2019) GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics 12(5):1–8
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS (2017) TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 77(21):e108–e110
Jézéquel P, Frénel J-S, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M (2013) bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database 2013
Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN (2020) Visualizing and interpreting cancer genomics data via the xena platform. Nat Biotechnol 38(6):675–678
Xie Z, Bailey A, Kuleshov MV, Clarke DJ, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM (2021) Gene set knowledge discovery with enrichr. Curr Protoc 1(3):e90
Győrffy B (2021) Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J 19:4101–4109
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A (2015) Tissue-based map of the human proteome. Science 347(6220):1260419
Lánczky A, Győrffy B (2021) Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res 23(7):e27633
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14(1):1–14
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44(W1):W90–W97
Kaur M, Mehta V, Wani AA, Arora S, Bharatam PV, Sharon A, Singh S, Kumar R (2021) Synthesis of 1, 4-dihydropyrazolo [4, 3-b] indoles via intramolecular C (sp2)-N bond formation involving nitrene insertion, DFT study and their anticancer assessment. Bioorg Chem 114:105114
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408
Kaur M, Mehta V, Arora S, Munshi A, Singh S, Kumar R (2021) Design, synthesis and biological evaluation of new 5-(2-Nitrophenyl)-1-aryl-1H-pyrazoles as Topoisomerase inhibitors. ChemistrySelect 6(26):6644–6651
Li X, Yang J, Peng L, Sahin AA, Huo L, Ward KC, O’Regan R, Torres MA, Meisel JL (2017) Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat 161(2):279–287
Hennigs A, Riedel F, Gondos A, Sinn P, Schirmacher P, Marmé F, Jäger D, Kauczor H-U, Stieber A, Lindel K (2016) Prognosis of breast cancer molecular subtypes in routine clinical care: a large prospective cohort study. BMC Cancer 16(1):1–9
Duffy MJ, Synnott NC, Crown J (2018) Mutant p53 in breast cancer: potential as a therapeutic target and biomarker. Breast Cancer Res Treat 170(2):213–219
Suman P, Mehta V, Craig AW, Chander H (2022) Wild type p53 suppresses formin binding protein 17 (FBP17) to reduce invasion. Carcinogenesis 43(5):494–503. https://doi.org/10.1093/carcin/bgac015
Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MD, Niu B, McLellan MD, Uzunangelov V (2014) Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158(4):929–944
Muller PA, Vousden KH (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25(3):304–317
Ding W, Chen G, Shi T (2019) Integrative analysis identifies potential DNA methylation biomarkers for pan-cancer diagnosis and prognosis. Epigenetics 14(1):67–80
Chander H, Truesdell P, Meens J, Craig AW (2013) Transducer of Cdc42-dependent actin assembly promotes breast cancer invasion and metastasis. Oncogene 32(25):3080–3090
Blanco MA, Kang Y (2011) Signaling pathways in breast cancer metastasis-novel insights from functional genomics. Breast Cancer Res 13(2):1–9
Hsu JL, Hung M-C (2016) The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev 35(4):575–588
Suman P, Mishra S, Chander H (2018) High expression of FBP17 in invasive breast cancer cells promotes invadopodia formation. Med Oncol 35(5):1–6
Mehta V, Chander H, Munshi A (2021) Mechanisms of anti-tumor activity of Withania somnifera (Ashwagandha). Nutr Cancer 73(6):914–926
Kumar A, Tikoo S, Maity S, Sengupta S, Sengupta S, Kaur A, Kumar Bachhawat A (2012) Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep 13(12):1095–1101
Reich S, Nguyen CD, Has C, Steltgens S, Soni H, Coman C, Freyberg M, Bichler A, Seifert N, Conrad D (2020) A multi-omics analysis reveals the unfolded protein response regulon and stress-induced resistance to folate-based antimetabolites. Nat Commun 11(1):1–15
Madden E, Logue SE, Healy SJ, Manie S, Samali A (2019) The role of the unfolded protein response in cancer progression: from oncogenesis to chemoresistance. Biol Cell 111(1):1–17
Nagelkerke A, Bussink J, Mujcic H, Wouters BG, Lehmann S, Sweep FC, Span PN (2013) Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res 15(1):1–13
Lin HH, Chung Y, Cheng C-T, Ouyang C, Fu Y, Kuo C-Y, Chi KK, Sadeghi M, Chu P, Kung H-J (2018) Autophagic reliance promotes metabolic reprogramming in oncogenic KRAS-driven tumorigenesis. Autophagy 14(9):1481–1498
Yang H, He X, Zheng Y, Feng W, Xia X, Yu X, Lin Z (2014) Down-regulation of asparagine synthetase induces cell cycle arrest and inhibits cell proliferation of breast cancer. Chem Biol Drug Des 84(5):578–584
Acknowledgements
The authors highly acknowledge the developers of various databases and web servers used in the study. The current research was in part supported by extramural research funding from the Department of Science and Technology-Science and Engineering Research Board (DST-SERB), India (Grant Nos. EMR/2015/000761 & EEQ/2017/000794). V.M. is thankful to ICMR, New Delhi for providing the Senior Research Fellowship (SRF).
Funding
The study was funded by the Department of Science and Technology-Science and Engineering Research Board (DST-SERB), India (Grant Nos. EMR/2015/000761 & EEQ/2017/000794).
Author information
Authors and Affiliations
Contributions
VM and HC: designed the study. VM: performed the experimental work. VM and HC: drafted the figures and prepared the manuscript. JM, HK, and AM: performed data analysis in part. All the authors analyzed the data and reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval and Informed consent
No ethical approval and consent were required for the current study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mehta, V., Meena, J., Kasana, H. et al. Prognostic significance of CHAC1 expression in breast cancer. Mol Biol Rep 49, 8517–8526 (2022). https://doi.org/10.1007/s11033-022-07673-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07673-x