Abstract
Background
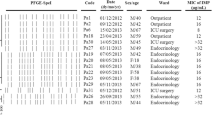
Understanding the mechanisms of antibiotic resistance is important for designing new therapeutic options and controlling resistant strains. The goal of this study was to look at the molecular epidemiology and mechanisms of resistance in carbapenem-resistant Pseudomonas aeruginosa (CRPA) isolates from Tabriz, Iran.
Methods
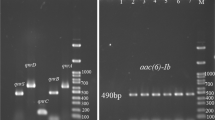
One hundred and forty P. aeruginosa were isolated and antibiotic susceptibility patterns were determined. Overproduction of AmpC and efflux pumps were discovered using phenotypic techniques. Polymerase chain reaction (PCR) was used to determine the presence of carbapenemase-encoding genes. In addition, the expressions of OprD and efflux pumps were evaluated by the Real-Time PCR. Random amplified polymorphic DNA typing (RAPD) was performed for genotyping.
Results
Among 140 P. aeruginosa isolates, 74 (52.8%) were screened as CRPA. Overexpression of efflux systems was observed in 81% of isolates, followed by decreased expression of OprD (62.2%), presence of carbapenemase genes (14.8%), and overproduction of AmpC (13.5%). In most isolates, carbapenem resistance was multifactorial (60.8%). According to our results, the prevalence of CRPA is at alarming levels. Overexpression of efflux systems was the most common mechanism of carbapenem resistance.
Conclusion
Most isolates may originate in patients themselves, but cross-infection is possible. Therefore, we suggest a pattern shift in the strategy of CRPA in our setting.


Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article [and its tables and figures].
References
Pachori P, Gothalwal R, Gandhi P (2019) Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis 6:109–119
Botelho J, Grosso F, Peixe L (2019) Antibiotic resistance in Pseudomonas aeruginosa—mechanisms, epidemiology and evolution. Drug Resist Updates 44:100640
Aghapour Z, Gholizadeh P, Ganbarov K, Bialvaei AZ, Mahmood SS, Tanomand A et al (2019) Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist 12:965–975
McCracken MG, Adam HJ, Blondeau JM, Walkty AJ, Karlowsky JA, Hoban DJ et al (2019) Characterization of carbapenem-resistant and XDR Pseudomonas aeruginosa in Canada: results of the CANWARD 2007–2016 study. J Antimicrob Chemother 74:iv32–iv38
Akhi MT, Khalili Y, Ghotaslou R, Yousefi S, Kafil HS, Naghili B et al (2018) Evaluation of carbapenem resistance mechanisms and its association with Pseudomonas aeruginosa infections in the Northwest of Iran. Microb Drug Resist 24:126–135
Zahedi bialvaei A, Rahbar M, Hamidi-Farahani R, Asgari A, Esmailkhani A, Mardani dashti Y et al (2021) Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb Pathogen 153:104789
Diene SM, L’homme T, Bellulo S, Stremler N, Dubus J-C, Mely L et al (2013) ISPa46, a novel insertion sequence in the oprD porin gene of an imipenem-resistant Pseudomonas aeruginosa isolate from a cystic fibrosis patient in Marseille, France. Int J Antimicrob Agents 42:268–271
Schäfer E, Malecki M, Tellez-Castillo CJ, Pfennigwerth N, Marlinghaus L, Higgins PG et al (2019) Molecular surveillance of carbapenemase-producing Pseudomonas aeruginosa at three medical centres in Cologne, Germany. Antimicrob Resist Infect Control 8:208
Neyestanaki DK, Mirsalehian A, Rezagholizadeh F, Jabalameli F, Taherikalani M, Emaneini M (2014) Determination of extended spectrum beta-lactamases, metallo-beta-lactamases and AmpC-beta-lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. Burns 40:1556–1561
Bahar MA, Jamali S, Samadikuchaksaraei A (2010) Imipenem-resistant Pseudomonas aeruginosa strains carry metallo-β-lactamase gene blaVIM in a level I Iranian burn hospital. Burns 36:826–830
Yousefi S, Farajnia S, Nahaei MR, Akhi MT, Ghotaslou R, Soroush MH et al (2010) Detection of metallo-β-lactamase–encoding genes among clinical isolates of Pseudomonas aeruginosa in northwest of Iran. Diagn Microbiol Infect Dis 68:322–325
Bialvaei AZ, Kafil HS, Leylabadlo HE, Asgharzadeh M, Aghazadeh M (2015) Dissemination of carbapenemases producing gram negative bacteria in the middle east. Iranian J Microbiol 7:226–246
CLSI (2017) M100-S27. Performance standards for antimicrobial susceptibility testing: 27th informational supplement. CLSI, Wayne
Wang M, Wei H, Zhao Y, Shang L, Di L, Lyu C et al (2019) Analysis of multidrug-resistant bacteria in 3223 patients with hospital-acquired infections (HAI) from a tertiary general hospital in China. Bosn J Basic Med Sci 19:86
Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C et al (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281
Rodríguez-Martínez J-M, Poirel L, Nordmann P (2009) Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:4783–4788
Akhi MT, Ghotaslou R, Alizadeh N, Yekani M, Beheshtirouy S, Asgharzadeh M et al (2017) nim gene-independent metronidazole-resistant Bacteroides fragilis in surgical site infections. GMS Hygiene Infect Control 12:Doc13
Poirel L, Walsh TR, Cuvillier V, Nordmann P (2011) Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123
Riera E, Cabot G, Mulet X, García-Castillo M, del Campo R, Juan C et al (2011) Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: impact on the activity of imipenem, meropenem and doripenem. J Antimicrob Chemother 66:2022–2027
Hematzadeh A, Haghkhah M (2021) Biotyping of isolates of Pseudomonas aeruginosa isolated from human infections by RAPD and ERIC-PCR. Heliyon 7:e07967
Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP (1996) Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol 34:1129–1135
Zamani K, Irajian G, Bialvaei AZ, Salehi TZ, Khormali M, Vosough A et al (2022) Passive immunization with anti-chimeric protein PilQ/PilA–DSL region IgY does not protect against mortality associated with Pseudomonas aeruginosa sepsis in a rabbit model. Mol Immunol 141:258–264
Khalili Y, Memar MY, Farajnia S, Adibkia K, Kafil HS, Ghotaslou R (2021) Molecular epidemiology and carbapenem resistance of Pseudomonas aeruginosa isolated from patients with burns. J Wound Care 30:135–141
Zahedi bialvaei A, Razavi S, Notash Haghighat F, Hemmati A, Akhavan MM, Jeddi-Tehrani M et al (2021) Monoclonal antibody directed to the PilQ -PilA DSL region in Pseudomonas aeruginosa improves survival of infected mice with antibiotic combination. Microb Pathogen 158:105060
Wang J, Zhou J-Y, Qu T-T, Shen P, Wei Z-Q, Yu Y-S et al (2010) Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Chinese hospitals. Int J Antimicrob Agents 35:486–491
Martis N, Leroy S, Blanc V (2014) Colistin in multi-drug resistant Pseudomonas aeruginosa blood-stream infections: a narrative review for the clinician. J Infect 69:1–12
Chatterjee M, Anju C, Biswas L, Kumar VA, Mohan CG, Biswas R (2016) Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int J Med Microbiol 306:48–58
Hu Y, Liu C, Wang Q, Zeng Y, Sun Q, Shu L et al (2021) Emergence and expansion of a carbapenem-resistant Pseudomonas aeruginosa clone are associated with plasmid-borne bla KPC-2 and virulence-related genes. Msystems 6:e00154-e221
Laupland KB, Parkins MD, Church DL, Gregson DB, Louie TJ, Conly JM et al (2005) Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of Metallo-β-Lactamase (MBL)—producing strains. J Infect Dis 192:1606–1612
Pournaras S, Maniati M, Spanakis N, Ikonomidis A, Tassios P, Tsakris A et al (2005) Spread of efflux pump-overexpressing, non-metallo-β-lactamase-producing, meropenem-resistant but ceftazidime-susceptible Pseudomonas aeruginosa in a region with bla VIM endemicity. J Antimicrob Chemother 56:761–764
Garvey M, Bradley C, Jumaa P (2016) Environmental decontamination following occupancy of a burns patient with multiple carbapenemase-producing organisms. J Hosp Infect 93:136–140
Bialvaei AZ, Samadi KH (2015) Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin 31:707–721
Rahimi E, Asgari A, Azimi T, Soleiman-Meigooni S (2021) Molecular detection of carbapenemases and extended-spectrum β-lactamases-encoding genes in clinical isolates of pseudomonas aeruginosa in Iran. Jundishapur J Microbiol 14:1–6
Saderi H, Lotfalipour H, Owlia P, Salimi H (2010) Detection of metallo-β-lactamase producing Pseudomonas aeruginosa isolated from burn patients in Tehran. Iran Lab Med 41:609–612
Kao C-Y, Chen S-S, Hung K-H, Wu H-M, Hsueh P-R, Yan J-J et al (2016) Overproduction of active efflux pump and variations of OprD dominate in imipenem-resistant Pseudomonas aeruginosa isolated from patients with bloodstream infections in Taiwan. BMC Microbiol 16:107
Quale J, Bratu S, Gupta J, Landman D (2006) Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 50:1633–1641
Wang W, Wang X (2020) Prevalence of metallo-β-lactamase genes among Pseudomonas aeruginosa isolated from various clinical samples in China. J Lab Med 44:197–203
Mansour W, Poirel L, Bettaieb D, Bouallegue O, Boujaafar N, Nordmann P (2009) Metallo-β-lactamase–producing Pseudomonas aeruginosa isolates in Tunisia. Diagn Microbiol Infect Dis 64:458–461
Khuntayaporn P, Montakantikul P, Santanirand P, Kiratisin P, Chomnawang MT (2013) Molecular investigation of carbapenem resistance among multidrug-resistant Pseudomonas aeruginosa isolated clinically in Thailand. Microbiol Immunol 57:170–178
Dantas RCC, e Silva RT, Ferreira ML, Gonçalves IR, Araújo BF, de Campos PA et al (2017) Molecular epidemiological survey of bacteremia by multidrug resistant Pseudomonas aeruginosa: the relevance of intrinsic resistance mechanisms. PLoS ONE 12:e0176774
Acknowledgements
We would appreciate the cooperation of microbiology laboratory personnel from hospitals of Tabriz in the collection of clinical isolates. We thank for dear Dr. Kamali chief of Aryan laboratory to worthy assistance in our work.
Funding
This project was financially supported by the Immunology Research Center, Tabriz University of Medical Sciences.
Author information
Authors and Affiliations
Contributions
YK: Conceptualization, study supervision, design of the study, and Writing- Original draft preparation. PO, HRG: Data curation, Investigation, Software, Validation. SZ, FB, AE; Visualization, Methodology, Project administration. AZB; Supervision, Methodology, Reviewing and Editing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the research ethics committee (TBZMED.REC.2015.161) in Tabriz University of Medical Sciences, Tabriz, Iran.
Consent to participate
The demographic data were obtained from the medical records of each patient with informed consent for all patients or parents.
Consent for publication
The participants have consented to the submission of this article to the journal. We confirm that the manuscript, or part of it, has neither been published nor is currently under consideration for publication. This work and the manuscript were approved by all co-authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khalili, Y., Omidnia, P., Goli, H.R. et al. Molecular characterization of carbapenem-resistant Pseudomonas aeruginosa isolated from four medical centres in Iran. Mol Biol Rep 49, 8281–8289 (2022). https://doi.org/10.1007/s11033-022-07640-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07640-6