Abstract
Background
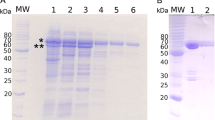
We have identified endogenous p65 to be an SDS-stable dimer protein composed of ~ 37 kDa hnRNPA/B-like subunits. We have investigated molecular properties involved in the stability of dimeric form, and their regulation in the transition between monomeric and dimeric forms of hnRNPA/B-like protein 2. We also investigated a cellular property conserved between squid hnRNPA/B-like protein 2 and human hnRNPA1 protein in a neuronal context.
Methods and results
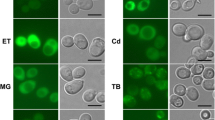
Here we show biochemical properties of a recombinant hnRNPA/B-like protein 2 (rP2) in vitro experiments, as one of p65 subunit. We found that interaction between rP2 and RNA molecules interfered with the dynamics of rP2 dimers formation, involved in disulfide bonds and/or postranslational alterations in distinct stage of SDS-stable dimers formation. In addition, we have performed immunofluorescence in SH-SY5Y cells and observed that the pEGFP-P2 fusion protein was expressed in the nucleus, similar to what is observed for human hnRNPA1 protein.
Conclusion
Our results reinforce the idea that p65 is an SDS-stable dimer. Thus, a deeper understanding between monomeric and dimeric transition dynamic is critical into evolution of several neurodegenerative disease.






Similar content being viewed by others
Data availability
All data generated in this study are available in the manuscript. This manuscript is published in the bioRxi as a preprint https://doi.org/10.1101/2021.07.02.450876
Abbreviations
- rP2:
-
Recombinant squid hnRNPA/B-like protein 2
- pEGFP-P2:
-
PEGFP-C1 vector containing the ORF of squid hnRNPA/B-like protein 2
- mCherry-P2:
-
PmCherry-C1 vectors containing the ORF of squid hnRNPA/B-like protein 2
- PABP-1:
-
Poly-A binding protein-1
- DND1:
-
Dead end protein homolog1
- hnRNP:
-
Heterogeneous nuclear ribonucleoprotein
- ORF:
-
Open reading frame
- PAGE:
-
Polyacrylamide gel electrophoresis
- PBS:
-
Phosphate buffered saline
- RNP:
-
Ribonucleoprotein
- RNP1/RNP2:
-
Core sequences of RNA recognition motifs
- SDS:
-
Sodium dodecyl sulfate
- SG:
-
Stress granules
- BFS:
-
Bovine fetal serum
- FRAP:
-
Fluorescence recovery after photobleach
References
Dreyfuss G, Kim VN, Kataoka N (2002) Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3:195–205
He Y, Smith R (2009) Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci 166(7):239–1256
McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GAV, Velde C (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet 20:1400–1410
Siomi H, Dreyfuss G (1995) A nuclear localization domain in the hnRNP A1 protein. J Cell Biol 129(3):551–560
Michael WM, Choi M, Dreyfuss G (1995) A nuclear export signal in hnRNP A1: a signal mediated, temperature-dependent nuclear protein export pathway. Cell 83(3):415–422
Lico DT, Lopes GS, Brusco J, Rosa JC, Gould RM, De Giorgis JA, Larson RE (2015) A novel SDS-stable dimer of a heterogeneous nuclear ribonucleoprotein at presynaptic terminals of squid neurons. Neuroscience 300:381–392
Lopes GS, Lico DTP, Rocha RS, Oliveira RR, Sebollela AS, Paco-Larson ML, Larson RE (2019) A phylogenetically conserved hnRNP type A/B protein from squid brain. Neurosci Lett 696:219–224
Gabriel LS, Janaina B, José RC, Roy LE, Diego LTP (2020) Selectively RNA interaction by a hnRNPA/B-like protein at presynaptic terminal of squid neuron. Invertebr Neurosci 20:1–14
Joshua DN, Gardner LA, Salapa HE, Levin MC (2016) Antibodies to the RNA binding protein heterogeneous nuclear ribonucleoprotein A1 colocalize to stress granules resulting in altered RNA and protein levels in a model of neurodegeneration in multiple sclerosis. J Clin Cell Immunol 7(2):402
George E, Wasiak S, Blandford V, Tong X-K, Serrano A, Fan X, del Rayo M, Sánchez-Carbente FS, Bell AW, Boismenu D, Lacaille J-C, McPherson PS, DesGroseillers L, Sossin WS (2006) Characterization of an RNA granule from developing. BrainMol Cell Proteomics 4:635–651
Purice MD, Taylor JP (2018) Linking hnRNP function to ALS and FTD pathology. Front Neurosci 12:326
Heraviab YB, Van Broeckhovenab C, der Zee J (2020) Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol Dis 134:104639
Guil S, Long JC, Cáceres JF (2006) hnRNPA1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol 26(15):5744–5758
Banani SF, Allyson M, Rice WB, Peeples YL, Jain S, Parker R, Rosen MK (2016) Compositional control of phase-separated cellular bodies. Cell 166:651–663
Protter DSW, Parker R (2016) Principles and properties of stress granules. Trends Cell Biol 26(9):668–679
Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, Taylor JP, Gitler AD, Rothstein JD, Lloyd TE (2018) Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173:958-971.e917
Polymenidou M, Cleveland DW (2017) Biological spectrum of amyotrophic lateral sclerosis prions. Cold Spring Harb Perspect Med 7:a024133
Ramaswami M, Taylor JP, Parker R (2013) Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154:727–736
Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R (2017) The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68:808-820.e5
Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, Myong S, Gladfelter AS (2018) mRNA structure determines specificity of a polyQ-driven phase separation. Science 360:922–927
d'Errico, Melanie Meyer-Luehmann (2020) Mechanisms of Pathogenic Tau and Aß Protein Spreading in Alzheimer's Disease. Front Aging Neurosci; 27;12:265.) (Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, Gu J, Xie M, Zhao K, Dai B, Shin WS, He J, He L, Jiang L, Zhao M, Sun B, Li X, Liu C, Li D. (2019) Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun 10(1):2006.)
Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillén-Boixet J, Franzmann TM, Jahnel M, Marrone L, Chang YT, Sterneckert J, Tomancak P, Hyman AA, Alberti S (2018) RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360:918–921
Buratti E (2018) TDP-43 post-translational modifications in health and disease. Expert Opin Ther Targets 22:279–293
Aguzzi A, Rajendran L (2009) The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 64(6):783–790
Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J et al (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1cause multisystem proteinopathy and ALS. Nature 495:467–473
Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH Jr (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323:1205–1208
Liu Q, Shu S, Wang RR, Liu F, Cui B, Guo XN, Lu CX, Li XG, Liu MS, Peng B, Cui LY, Zhang X (2016) Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology 87:1763–1769
Liu YC, Chiang PM, Tsai KJ (2013) Disease animal models of TDP-43 proteinopathy and their pre-clinical applications. Int J Mol Sci 14:20079–20111
Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, Matchett BJ, Mittag T, Temirov J, Hsiung GR, Krieger C, Murray ME, Kato M, Fryer JD, Petrucelli L, Zinman L, Weintraub S, Mesulam M, Keith J, Zivkovic SA, Hirsch-Reinshagen V, Roos RP, Zuchner S, Graff-Radford NR, Petersen RC, Caselli RJ, Wszolek ZK, Finger E, Lippa C, Lacomis D, Stewart H, Dickson DW, Kim HJ, Rogaeva E, Bigio E, Boylan KB, Taylor JP, Rademakers R (2017) TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95:808-816.e809
Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R (2018) Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep 22:1401–1412
Lico DTP, Rosa JC, DeGiorgis JA, deVasconcelos EJR, Casaletti L, Tauhata SBF, Baqui MMA, Fukuda M, Moreira JE, Larson RE (2010) A novel 65 kDa RNA-binding protein in squid presynaptic terminals. Neuroscience 166:73–83
Chomczynski P, Sacchi N (1987) Single-step method of RNA acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
McLellan T (1982) Electrophoresis buffers for polyacrylamide gels at various pH. Anal Biochem 126:94–99
Kedersha N, Ivanov P, Anderson P (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci 38:494–506
Kedersha NL, Gupta M, Li W, Miller I, Anderson P (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147:1431–1442
Lindquist S (1981) Regulation of protein synthesis during heat shock. Nature 293:311–314
Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R (2016) Distinct stages in stress granule assembly and disassembly. Elife Trends Biochem Sci 38:494–506
Christopher FL, Lentz A, Mayfield SP (2000) Disulfide bond formation between RNA binding domains is used to regulate mRNA binding activity of the chloroplast poly(A)-binding protein. J Biol Chem 275:8275–8278
Kumari P, Bhavesh NS (2021) Human DND1-RRM2 forms a non-canonical domain swapped dimer. Protein Sci 6:1184–1195
Levengood JD, Tolbert BS (2018) Idiosyncrasies of hnRNP A1-RNA recognition: can binding mode influence function. Semin Cell Dev Biol 86:150–161
Liu X, Ishizuka T, Bao H-L, Wada K, Takeda Y, Iida K et al (2017) Structure-dependent binding of hnRNPA1 to telomere RNA. J Am Chem Soc 139:7533–7539
Cieniková Z, Jayne S, Damberger FF, Allain FH-T, Maris C (2015) Evidence for cooperative tandem binding of hnRNP C RRMs in mRNA processing. RNA 21:1931–1942
Gagné M, Deshaies J-E, Sidibé H, Benchaar Y, Arbour D, Dubinski A, Litt G, Peyrard S, Robitaille R, Sephton CF, Vande Velde C (2021) hnRNP A1B, a splice variant of HNRNPA1, is spatially and temporally regulated. Front Neurosci 15:724307
Afroz T, Hock E-M, Ernst P, Foglieni C, Jambeau M, Gilhespy LAB, Laferriere F, Maniecka Z, Plückthun A, Mittl P, Paganetti P, Allain FHT, Polymenidou M (2017) Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat Commun 8(1):45
Rengifo-Gonzalez JC, El Hage K, Clément MJ, Steiner E, Joshi V, Craveur P, Durand D, Pastré D, Bouhss A (2021) The cooperative binding of TDP-43 to GU-rich RNA repeats antagonizes TDP-43 aggregation. Elife 10:e67605
Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, Shewmaker F (2018) A single N-terminal hosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J 37(5):e97452
Aguero T, Jin Z, Owens D, Malhotra A, Newman K, Yang J, Lou KM (2018) Combined functions of two RRMs in dead-end1 mimic helicase activity to promote nanos1 translation in the germline. Mol Reprod Dev 85:896–908
Maris C, Dominguez C, Allain FH (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272:2118–2131
Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163:123–133
Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R (2018) RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci USA 115:2734–2739
Acknowledgements
Thanks to Silvia Andrade and Domingos Pitta for expert technical help, and M.Sc Elizabete R. Milani for technical help with confocal microscopy, performed at Laboratório Multiusuário de Microscopia Confocal—LMMC, Fapesp 2004/08868-0. Special thanks to Lexie Friend, PhD, Honorary Fellow School of Chemistry and Molecular Biosciences at the University of Queensland St Lucia, 4072, Australia. She did make suggestions important to the manuscript and my works that it supported my finds in discussion section
Funding
The Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); no specifc grant was received for this Publisher.
Author information
Authors and Affiliations
Contributions
DTPL (performed experiments biochemical and prepared manuscript); GSL (performed experiments immunofluorescence); MLPL (analyzed data) and REL (designed the experiments). and Contributions Structural the Marine Resources Center of the Marine Biological Laboratory in Woods Hole, MA and f the Centro de Biologia Marinha-CEBIMar, University of São Paulo, São Sebastião, Brazil.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of conflict of interest.
Ethical approval
The number of animals used has been conformed to ethical guidelines presented by National System of Nature Conservation Units Snuc-Ibama/Brazil with solicitation Nº 53615US and Public Health Service, National Institutes of Health Publication according to Law No. 9,985/2000.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lopes, G.S., Lico, D.T.P. Biochemical and subcellular characterization of a squid hnRNPA/B-like protein 2 in osmotic stress activated cells reflects molecular properties conserved in this protein family. Mol Biol Rep 49, 4257–4268 (2022). https://doi.org/10.1007/s11033-022-07260-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07260-0