Abstract
Background
The selection and validation of stably expressed reference genes is key for accurately quantifying the mRNA abundance of genes under different treatments. In the rabbit model of fasting caecotrophy, reports about the selection of stable reference genes are not available.
Methods and results
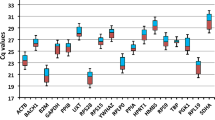
This study aims to screen suitable reference genes in different tissues (including uterus, cecum, and liver) of rabbits between control and fasting caecotrophy groups. RT-qPCR was used to analyze the expression levels of eight commonly used reference genes (including GAPDH, 18S rRNA, B2M, CYP, HPRT1, β-actin, H2afz, Ywhaz), and RefFinder (including geNorm, NormFinder, and BestKeeper) was used to analyze the expression stability of these reference genes. Our results showed that the most stable reference genes were different in different tissues and treatments. In the control and fasting caecotrophy groups, CYP, GAPDH and HPRT1 were proven to be the top stable reference genes in the uterus, cecum, and liver tissues, respectively. GAPDH and Ywhaz were proven to be the top two stable reference genes among uterus, cecum, and liver in both control and fasting caecotrophy groups.
Conclusions
Our results indicated that the combined analysis of three or more reference genes (GAPDH, HPRT1, and Ywhaz) are recommended to be used for RT-qPCR normalization in the rabbit model of fasting caecotrophy, and that GAPDH is a better choice than the other reference genes for normalizing the relative expression of target genes in different tissues of fasting caecotrophy rabbits.



Similar content being viewed by others
Data availability
All data are fully available without restriction.
References
Kuijper DPJ, van Wieren SE, Bakker JP (2004) Digestive strategies in two sympatrically occurring lagomorphs. J Zool 264:171–178. https://doi.org/10.1017/S0952836904005722
Wang Y, Xu H, Sun G et al (2019) Transcriptome analysis of the effects of fasting caecotrophy on hepatic lipid metabolism in New Zealand rabbits. Animals (Basel). https://doi.org/10.3390/ani9090648
Bo T-B, Zhang X-Y, Kohl KD et al (2020) Coprophagy prevention alters microbiome, metabolism, neurochemistry, and cognitive behavior in a small mammal. ISME J 14:2625–2645. https://doi.org/10.1038/s41396-020-0711-6
Li R, Li X, Huang T et al (2020) Influence of cecotrophy on fat metabolism mediated by caecal microorganisms in New Zealand white rabbits. J Anim Physiol Anim Nutr (Berl) 104:749–757. https://doi.org/10.1111/jpn.13309
Bustin SA, Nolan T (2004) Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 15:155–166
Imbeaud S, Graudens E, Boulanger V et al (2005) Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res 33:e56. https://doi.org/10.1093/nar/gni054
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284. https://doi.org/10.1038/sj.gene.6364190
Ho-Pun-Cheung A, Cellier D, Lopez-Crapez E (2008) Considerations for normalisation of RT-qPCR in oncology. Ann Biol Clin (Paris) 66:121–129. https://doi.org/10.1684/abc.2008.0204
Toorani T, Mackie PM, Mastromonaco GF (2021) Validation of reference genes for use in untreated bovine fibroblasts. Sci Rep 11:10253. https://doi.org/10.1038/s41598-021-89657-8
Cai C, Cai P, Chu G (2019) Selection of suitable reference genes for core clock gene expression analysis by real-time qPCR in rat ovary granulosa cells. Mol Biol Rep 46:2941–2946. https://doi.org/10.1007/s11033-019-04755-1
Chen J, Bao Z, Huang Y et al (2020) Selection of suitable reference genes for qPCR gene expression analysis of HepG2 and L02 in four different liver cell injured models. Biomed Res Int 2020:8926120. https://doi.org/10.1155/2020/8926120
Mamo S, Gal AB, Polgar Z, Dinnyes A (2008) Expression profiles of the pluripotency marker gene POU5F1 and validation of reference genes in rabbit oocytes and preimplantation stage embryos. BMC Mol Biol 9:67. https://doi.org/10.1186/1471-2199-9-67
Nachar W, Busseuil D, Shi Y et al (2014) Optimisation of reference genes for gene-expression analysis in a rabbit model of left ventricular diastolic dysfunction. PLoS One 9:e89331. https://doi.org/10.1371/journal.pone.0089331
Pfaffl Michael W, Tichopad Ales, Prgomet Christian, Neuvians Tanja P (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–excel-based tool using pair-wise correlations. Biotechnol Lett. https://doi.org/10.1023/b:bile.0000019559.84305.47
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496
Vandesompele J, De Preter K, Pattyn F et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034
Xie F, Xiao P, Chen D et al (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. https://doi.org/10.1007/s11103-012-9885-2
Klenke S, Renckhoff K, Engler A et al (2016) Easy-to-use strategy for reference gene selection in quantitative real-time PCR experiments. Naunyn Schmiedebergs Arch Pharmacol 389:1353–1366. https://doi.org/10.1007/s00210-016-1305-8
Kozera B, Rapacz M (2013) Reference genes in real-time PCR. J Appl Genet 54:391–406. https://doi.org/10.1007/s13353-013-0173-x
Derveaux S, Vandesompele J, Hellemans J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50:227–230. https://doi.org/10.1016/j.ymeth.2009.11.001
Bustin SA, Beaulieu J-F, Huggett J et al (2010) MIQE précis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol 11:74. https://doi.org/10.1186/1471-2199-11-74
Sandercock DA, Coe JE, Di Giminiani P, Edwards SA (2017) Determination of stable reference genes for RT-qPCR expression data in mechanistic pain studies on pig dorsal root ganglia and spinal cord. Res Vet Sci 114:493–501. https://doi.org/10.1016/j.rvsc.2017.09.025
Adeola F (2018) Normalization of gene expression by quantitative RT-PCR in human cell line: comparison of 12 endogenous reference genes. Ethiop J Health Sci 28:741–748. https://doi.org/10.4314/ejhs.v28i6.9
Mamo S, Gal AB, Bodo S, Dinnyes A (2007) Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev Biol 7:14. https://doi.org/10.1186/1471-213X-7-14
Dang W, Zhang X, Ma Q et al (2020) Selection of reference genes suitable for normalization of RT-qPCR data in glioma stem cells. Biotechniques 68:130–137. https://doi.org/10.2144/btn-2019-0098
Feng X, Xiong Y, Qian H et al (2010) Selection of reference genes for gene expression studies in porcine skeletal muscle using SYBR green qPCR. J Biotechnol 150:288–293. https://doi.org/10.1016/j.jbiotec.2010.09.949
Peletto S, Bertuzzi S, Campanella C et al (2011) Evaluation of internal reference genes for quantitative expression analysis by real-time PCR in ovine whole blood. Int J Mol Sci 12:7732–7747. https://doi.org/10.3390/ijms12117732
Jarczak J, Kaba J, Bagnicka E (2014) The validation of housekeeping genes as a reference in quantitative real time PCR analysis: application in the milk somatic cells and frozen whole blood of goats infected with Caprine arthritis encephalitis virus. Gene 549:280–285. https://doi.org/10.1016/j.gene.2014.07.063
Ferrara N, Adamis AP (2016) Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov 15:385–403. https://doi.org/10.1038/nrd.2015.17
Itoh N (2016) FGF10: A multifunctional mesenchymal-epithelial signaling growth factor in development, health, and disease. Cytokine Growth Factor Rev 28:63–69. https://doi.org/10.1016/j.cytogfr.2015.10.001
Funding
This research was jointly supported by the “National Key Research and Development Program of China (2018YFD0502203)” and “the Special Fund for Henan Agriculture Research System (S2013-08-G01)”.
Author information
Authors and Affiliations
Contributions
onceptualization, Huifen Xu, Ming Li and Hui He; funding acquisition, Ming Li; supervision, Mengke Ni; formal analysis, Shanshan Xing; validation, Zhichao Li and Lei Yu; writing-original draft, Hui He and Zhichao Li; writing-review and editing, Huifen Xu and Dehu Zhuo. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Ethical approval
This study was designed and performed according to the guidelines of the institutional Animal Care and Use Committee (IACUC) of College of Animal Science and technology of Henan Agricultural University, China (No: 11-0085).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, H., Li, Z., Ni, M. et al. Screening and stability analysis of reference genes in fasting caecotrophy model in rabbits. Mol Biol Rep 49, 1057–1065 (2022). https://doi.org/10.1007/s11033-021-06927-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06927-4