Abstract
Background
Hair is a frequently encountered biological evidence in personal identification. The amount of nuclear DNA that can be extracted from a single strand of rootless hair is most limited, making the detection of short tandem repeat (STR) polymorphisms difficult. To overcome these limitations, deletion/insertion polymorphisms (DIP) as a new type of genetic marker have shown their benefits in detecting low-copy-number DNA. The Investigator DIPplex kit contains 30 biallelic autosomal DIP and amelogenin. The analysis of DIPs combines the advantages of both STR and single nucleotide polymorphism analyses. Thus, this study aimed to detect the DIP distribution of individual hair shafts from individuals.
Methods and results
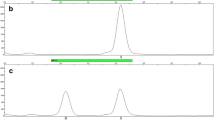
DNA was extracted from the shaft of fresh, aged, and shed hair. After DNA was evaluated, the DIP profiles were detected by capillary electrophoresis. The results indicated that the amount of DNA extracted from hair roots was much higher than that from the hair shafts in the same individual for all samples. The degradation index values of DNA from the aged hair shafts were highest. It is classified to be “mildly degraded.” Compared with their hair roots, the full DIP profiles were detected for fresh hair, 70% for aged hair, and 92% for shed hair. Contrarily, except for fresh hair shafts, only three STR loci of the aged and shed strands of hair could be genotyped using AmpFlSTR MiniFiler PCR Amplification Kit.
Conclusions
These results indicate that the detection of DIP profile is an effective tool for personal identification from hair shafts, including aged hair.




Similar content being viewed by others
Data availability
Data produced during this study are available from the corresponding author upon reasonable request. No specific materials were produced during this study.
References
Müller K, Klein R, Miltner E et al (2007) Improved STR typing of telogen hair root and hair shaft DNA. Electrophoresis 28(16):2835–2842
Brandhagen DM, Loreille O, Irwin JA (2018) Fragmented nuclear DNA is the predominant genetic material in human hair shafts. Genes (Basel) 9(12):640–660
Ottens R, Taylor D, Abarno D et al (2013) Successful direct amplification of nuclear markers from a single hair follicle. Forensic Sci Med Pathol 9:238–243
Parson W, Huber G, Moreno L et al (2015) Massively parallel sequencing of complete mitochondrial genomes from hair shaft samples. Forensic Sci Int Genet 15:8–15
Luce C, Montpetit S, Gangitano D et al (2009) Validation of the AmpFlSTR MiniFiler PCR amplification kit for use in forensic casework. J Forensic Sci 54(5):1046–1054
Grisedale KS, Murphy GM, Brown H et al (2018) Successful nuclear DNA profiling of rootless hair shafts: a novel approach. Int J Legal Med 132(1):107–115
Oldoni E, Podini D (2019) Forensic molecular biomarkers for mixture analysis. Forensic Sci Int Genet 41:107–119
Investigator DIPplex Handbook (2014), pp 6
Fondevila M, Phillips C, Santos C et al (2012) Forensic performance of two insertion-deletion marker assays. Int J Legal Med 126(5):725–737
QIAamp® DNA Investigator Handbook (2020), pp 39–43
Quantifiler TM HP and Trio DNA Quantification Kits, user guide (2018), pp 27–32
Bengtsson CE, Olsen ME, Brandt LØ et al (2012) DNA from keratinous tissue. Part I: hair and nail. Ann Anat 194(1):17–25
Mulero JJ, Chang CW, Lagacé RE et al (2008) Development and validation of the AmpFlSTR MiniFiler PCR amplification kit: a MiniSTR multiplex for the analysis of degraded and/or PCR inhibited DNA. J Forensic Sci 53(4):838–852
Horsman-Hall KM, Orihuela Y, Karczynski SL et al (2009) Development of STR profiles from firearms and fired cartridge cases. Forensic Sci Int Genet 3(4):242–250
Brandhagen MD, Loreille O, Irwin J (2018) Fragmented nuclear DNA is the predominant genetic material in human hair shafts. Genes 9(640):1–20
Klein R, Neumann C, Roy R (2015) Detection of insertion/deletion polymorphisme from challenged samples using Investigator DIPplex Kit. Forensic Sci Int Genet 16:29–37
Taberlet P, Griffin S, Goossens B et al (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 24(16):3189–3194
Whitaker JP, Cotton EA, Gill P (2001) A comparison of the characteristics of profiles produced with the AmpFlSTR SGM Plus multiplex system for both standard and low copy number STR DNA analysis. Forensic Sci Int 123(2–3):215–223
Acknowledgements
We would like to extend our thanks to Dr. Hirofumi Tsutsumi for his comments on the DNA evaluation of the samples.
Funding
This work was supported by Japan Society for the Promotion of Science KAKENHI, Great-in-Aid for Scientific Research (C) Number 21K10533.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This study was approved by the ethical committee of the Nihon University in accordance with ethical guidelines. (Decision No: 239-0). All procedures performed in studies involving human participants were in accordance with the ethical standards of this Ethics Committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all participants included in the study.
Consent to participate
All the authors listed have approved the manuscript that is enclosed.
Consent for publication
The manuscript is approved by all authors for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tie, J., Uchigasaki, S., Isobe, E. et al. Detection of deletion/insertion polymorphism profiles from single human hair shafts. Mol Biol Rep 49, 1017–1025 (2022). https://doi.org/10.1007/s11033-021-06921-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06921-w