Abstract
Background
High-throughput sequencing involves library preparation and amplification steps, which may induce contamination across samples or between samples and the environment.
Methods
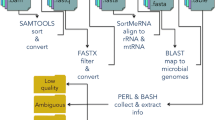
We tested the effect of applying an inline-index strategy, in which DNA indices of 6 bp were added to both ends of the inserts at the ligation step of library prep for resolving the data contamination problem.
Results
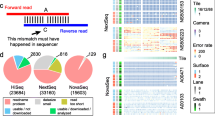
Our results showed that the contamination ranged from 0.29 to 1.25% in one experiment and from 0.83 to 27.01% in the other. We also found that contamination could be environmental or from reagents besides cross-contamination between samples.
Conclusions
Inline-index method is a useful experimental design to clean up the data and address the contamination problem which has been plaguing high-throughput sequencing data in many applications.





Similar content being viewed by others
Data availability
The raw reads have been uploaded to NCBI under Bioproject Numbers PRJNA749868, PRJNA750567.
Code availability
The custom codes can be found in online version of this article.
References
Eisenhofer R et al (2019) Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol 27(2):105–117
Laurin-Lemay S, Brinkmann H, Philippe H (2012) Origin of land plants revisited in the light of sequence contamination and missing data. Curr Biol 22(15):R593–R594
Sepulveda AJ et al (2020) The elephant in the lab (and Field): contamination in aquatic environmental DNA studies. Front Ecol Evol. https://doi.org/10.3389/fevo.2020.609973
Ballenghien M, Faivre N, Galtier N (2017) Patterns of cross-contamination in a multispecies population genomic project: detection, quantification, impact, and solutions. BMC Biol 15(1):25
Koutsovoulos G et al (2016) No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc Natl Acad Sci 113(18):5053–5058
Philippe H et al (2011) Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol 9(3):e1000602
Allio R et al (2020) Whole genome shotgun phylogenomics resolves the pattern and timing of swallowtail butterfly evolution. Syst Biol 69(1):38–60
Perez-Muñoz ME et al (2017) A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5(1):48
Racimo F, Renaud G, Slatkin M (2016) Joint estimation of contamination, error and demography for nuclear DNA from ancient humans. PLoS Genet 12(4):e1005972
Hänfling B et al (2016) Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol Ecol 25(13):3101–3119
Sepulveda AJ et al (2019) Adding invasive species biosurveillance to the U.S. Geological Survey streamgage network. Ecosphere 10(8):e02843
Serrao NR, Reid SM, Wilson CC (2018) Establishing detection thresholds for environmental DNA using receiver operator characteristic (ROC) curves. Conserv Genet Resour 10(3):555–562
Glassing A et al (2016) Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathogens 8(1):24
Dickins B et al (2014) Controlling for contamination in re-sequencing studies with a reproducible web-based phylogenetic approach. Biotechniques 56(3):134–141
Goldberg CS et al (2016) Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol Evol 7(11):1299–1307
De Simone G et al (2020) Contaminations in (meta) genome data: an open issue for the scientific community. IUBMB Life 72(4):698–705
Low AJ et al (2019) ConFindr: rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ 7:e6995
Straube N et al (2018) A phylogenomic approach to reconstruct interrelationships of main clupeocephalan lineages with a critical discussion of morphological apomorphies. BMC Evol Biol 18(1):158
Flickinger M et al (2015) Correcting for sample contamination in genotype calling of DNA sequence data. Am J Hum Genet 97(2):284–290
Rohland N et al (2015) Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos Trans R Soc Lond B Biol Sci 370(1660):20130624
Wang CC et al (2019) Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat Commun 10(1):590
Lipson M et al (2018) Population turnover in remote Oceania shortly after initial settlement. Curr Biol 28(7):1157-1165 e7
Meyer M, Kircher M (2010) Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. https://doi.org/10.1101/pdb.prot5448
Lin M, Zhang S, Yao M (2019) Effective detection of environmental DNA from the invasive American bullfrog. Biol Invasions 21(7):2255–2268
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461
Simion P et al (2018) A software tool ‘CroCo’ detects pervasive cross-species contamination in next generation sequencing data. BMC Biol 16(1):28
Kircher M, Sawyer S, Meyer M (2012) Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40(1):e3
Peyrégne S, Prüfer K (2020) Present-day DNA contamination in ancient DNA datasets. BioEssays 42(9):e2000081
Acknowledgements
We are grateful to Ms. Lifang Peng for helping with collecting water samples.
Funding
This work was supported by “Science and Technology Commission of Shanghai Municipality (19050501900)” and “Shanghai Academy of Environmental Sciences”.
Author information
Authors and Affiliations
Contributions
YW and CL conceived the research plan. YW and JH did the experiments and collected data. YW, HY and JH analyzed the data. YW and CL wrote the draft. All authors edited and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
CL, YW and HY are authors on a patent applied for by Shanghai Ocean University that covers the inline index technology (201811406204.X), and the remaining author (JH) has no financial conflict of interest.
Ethical approval
We followed the guidelines approved by the Ethical Committee of Shanghai Ocean University, China.
Consent to participate
All authors consent to participate.
Consent for publication
All authors consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., Yuan, H., Huang, J. et al. Inline index helped in cleaning up data contamination generated during library preparation and the subsequent steps. Mol Biol Rep 49, 385–392 (2022). https://doi.org/10.1007/s11033-021-06884-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06884-y