Abstract
Background
Recent investigations suggested that deregulated levels of Circular RNAs (circRNAs) could be associated with type 2 diabetes mellitus (T2DM) pathogenesis. Accordingly, this study aimed to determine the expression levels of circulating CircHIPK3, CDR1as and their correlation with biochemical parameters in patients with T2DM, pre-diabetes and control subjects.
Methods and results
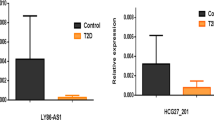
The expression of circRNAs in peripheral blood was determined using QRT-PCR in 70 patients with T2DM, 60 pre-diabetes and in 69 age and sex matched healthy controls. Moreover, bioinformatics tools were applied to explore and predict the potential interactions between circRNAs and other non-coding RNAs (ncRNAs). Our analysis revealed that the expression level of CircHIPK3 was significantly elevated in T2DM patients compared to healthy participants (P < 0.001) and pre-diabetes subjects (P = 0.018). In addition, ROC analysis suggested that at the cutoff value of 0.24 and the sensitivity and specificity of 50% and 88.4%, respectively, CircHIPK3 could distinguish between T2DM patients and control subjects. Furthermore, it was observed that the expression level of CDR1as is higher in pre-diabetic individuals than healthy individuals (P = 0.004). Finally, Spearman correlation analysis showed that there was a significant correlation between CircHIPK3 and CDR1as expression levels and clinical and anthropometrical parameters such as BMI, systolic and diastolic blood pressure, HbA1c and fasting blood glucose (P < 0.005).
Conclusions
The data of this study provided evidence that the expression levels of CircHIPK3, CDR1as increased in T2DM and pre-diabetes subjects, respectively.





Similar content being viewed by others
References
Tremblay J, Hamet P (2019) Environmental and genetic contributions to diabetes. Metabolism 100:153952
Saeedi P, Salpea P, Karuranga S, Petersohn I, Malanda B, Gregg EW et al (2020) Mortality attributable to diabetes in 20–79 years old adults, 2019 estimates: results from the International Diabetes Federation Diabetes Atlas. Diabetes Res Clin Pract 162:108086
Maass PG, Luft FC, Bähring S (2014) Long non-coding RNA in health and disease. J Mol Med 92(4):337–346
Cheng Y, Zhao P, Wu S, Yang T, Chen Y, Zhang X et al (2018) Cisplatin and curcumin co-loaded nano-liposomes for the treatment of hepatocellular carcinoma. Int J Pharm 545(1–2):261–273
Chen L-L (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17(4):205
Awan AR, Manfredo A, Pleiss JA (2013) Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans. Proc Natl Acad Sci 110(31):12762–12767
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci 73(11):3852–3856
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256
Hentze MW, Preiss T (2013) Circular RNAs: splicing’s enigma variations. EMBO J 32(7):923–925
Haque S, Harries LW (2017) Circular RNAs (circRNAs) in health and disease. Genes 8(12):353
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J (2013) Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 4:283
Ghasemi H, Sabati Z, Ghaedi H, Salehi Z, Alipoor B (2019) Circular RNAs in β-cell function and type 2 diabetes-related complications: a potential diagnostic and therapeutic approach. Mol Biol Rep 46:5631–5643
Xu H, Guo S, Li W, Yu P (2015) The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 5(1):1–12
Stoll L, Sobel J, Rodriguez-Trejo A, Guay C, Lee K, Venø MT et al (2018) Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab 9:69–83
Cai H, Jiang Z, Yang X, Lin J, Cai Q, Li X (2020) Circular RNA HIPK3 contributes to hyperglycemia and insulin homeostasis by sponging miR-192-5p and upregulating transcription factor forkhead box O1. Endocr J. https://doi.org/10.1507/endocrj.EJ19-0271
Wang L, Luo T, Bao Z, Li Y, Bu W (2018) Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochem Biophys Res Commun 505(3):644–650
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42
Meng X, Hu D, Zhang P, Chen Q, Chen M (2019) CircFunBase: a database for functional circular RNAs. Database. https://doi.org/10.1093/database/baz003
Shan K, Liu C, Liu B-H, Chen X, Dong R, Liu X et al (2017) Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 136(17):1629–1642
Sun Y, Chen R, Lin S, Xie X, Ye H, Zheng F et al (2019) Association of circular RNAs and environmental risk factors with coronary heart disease. BMC Cardiovasc Disord 19(1):223
Bazan HA, Hatfield SA, Brug A, Brooks AJ, Lightell DJ Jr, Woods TC (2017) Carotid plaque rupture is accompanied by an increase in the ratio of serum circR-284 to miR-221 levels. Circ: Cardiovasc Genet 10(4):e001720
Liu R, Zhang M, Ge Y (2020) Circular RNA HIPK3 exacerbates diabetic nephropathy and promotes proliferation by sponging miR-185. Gene 765:145065
Baker MA, Wang F, Liu Y, Kriegel AJ, Geurts AM, Usa K et al (2019) MiR-192-5p in the kidney protects against the development of hypertension. Hypertension 73(2):399–406
Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S et al (2016) Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep 6:22384
Li P, Yang X, Yuan W, Yang C, Zhang X, Han J et al (2018) CircRNA-Cdr1as exerts anti-oncogenic functions in bladder cancer by sponging MicroRNA-135a. Cell Physiol Biochem 46(4):1606–1616
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Qing S, Yuan S, Yun C, Hui H, Mao P, Wen F et al (2014) Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell Physiol Biochem 34(5):1733–1740
Sun Q, Yang Q, Xu H, Xue J, Chen C, Yang X et al (2019) miR-149 negative regulation of mafA is involved in the arsenite-induced dysfunction of insulin synthesis and secretion in pancreatic beta cells. Toxicol Sci 167(1):116–125
Wang S, Wen X, Han XR, Wang YJ, Shen M, Fan SH et al (2018) Repression of micro RNA-382 inhibits glomerular mesangial cell proliferation and extracellular matrix accumulation via FoxO1 in mice with diabetic nephropathy. Cell Prolif 51(5):e12462
Chen G, Lu L, Liu C, Shan L, Yuan D (2015) MicroRNA-377 suppresses cell proliferation and invasion by inhibiting TIAM1 expression in hepatocellular carcinoma. PLoS ONE 10(3):e0117714
Sekar D, Venugopal B, Sekar P, Ramalingam K (2016) Role of microRNA 21 in diabetes and associated/related diseases. Gene 582(1):14–18
Mazzeo A, Beltramo E, Lopatina T, Gai C, Trento M, Porta M (2018) Molecular and functional characterization of circulating extracellular vesicles from diabetic patients with and without retinopathy and healthy subjects. Exp Eye Res 176:69–77
Tang X, Miao Y, Luo Y, Sriram K, Qi Z, Lin F-M et al (2020) Suppression of endothelial AGO1 promotes adipose tissue browning and improves metabolic dysfunction. Circulation 142:365–379
Zhang C, Seo J, Murakami K, Salem ES, Bernhard E, Borra VJ et al (2018) Hepatic Ago2-mediated RNA silencing controls energy metabolism linked to AMPK activation and obesity-associated pathophysiology. Nat Commun 9(1):1–15
Huang Q, Yin J-Y, Dai X-P, Pei Q, Dong M, Zhou Z-G et al (2010) IGF2BP2 variations influence repaglinide response and risk of type 2 diabetes in Chinese population. Acta Pharmacol Sin 31(6):709–717
Chistiakov DA, Nikitin AG, Smetanina SA, Bel’chikova LN, Suplotova LA, Shestakova MV et al (2012) The rs11705701 G> A polymorphism of IGF2BP2 is associated with IGF2BP2 mRNA and protein levels in the visceral adipose tissue-a link to type 2 diabetes susceptibility. Rev Diabetic Stud: RDS 9(2–3):112
Funding
This work was financially supported by a Grant (960416) from the Deputy of Research, Yasuj University of Medical Sciences.
Author information
Authors and Affiliations
Contributions
BA planned the studies; BA and AM conducted analysis and interpretation of all experiments; FR and BK conducted all experiments. BA and GS wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
The study was approved by the Yasuj University of Medical Sciences Ethics Committee (IR.YUMS.REC.1397.153) and performed in compliance with the 1964 Helsinki declaration. Moreover, the informed consent was obtained from all subjects before enrolling in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rezaeinejad, F., Mirzaei, A., Khalvati, B. et al. Circulating expression levels of CircHIPK3 and CDR1as circular-RNAs in type 2 diabetes patients. Mol Biol Rep 49, 131–138 (2022). https://doi.org/10.1007/s11033-021-06850-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06850-8