Abstract
Background
Cryptochrome 1 (cry1), the core regulator of the circadian clock, is essential for ontogeny and mammalian reproduction. Unlike in other tissues, the cry1 gene have noncircadian functions in spermatogenesis, which implies the unique role of cry1 gene in the development of testis. The role of cry1 during the puberty has not been described yet. This study aimed to explore the relationship between cry1 expression and spermatogenic cell numbers.
Methods and results
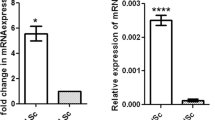
We analyzed testicular tissues from Hu sheep aged 0–180 days by hematoxylin and eosin staining, measured cry1 and cell proliferation regulatory factors (bricd5, tnfrsf21, cdk1) expression by quantitative real-time PCR and characterized the transcription factor in the 5′ flanking region of cry1 gene. The data revealed that the number of spermatocytes and early spermatocytes increased rapidly from 90 to 120 dpp (day postpartum). Correspondingly, there was a marked variation in the cry1 and cell proliferation related genes (bricd5, tnfrsf21, cdk1) mRNA expression in the testes from the age of 90 days to 180 days (p < 0.05). We also identified some transcription factors (tcfl5) related to cell proliferation.
Conclusions
There is a significant causal relationship between the transcription level of cry1 gene in Hu sheep testes and the number of spermatogenic cells. It is speculated that cry1 gene may regulate the proliferation of spermatogenic cells by regulating the expression of cell proliferation related genes such as bricd5, tnfrsf21 and cdk1.



Similar content being viewed by others
References
Moulla F, El-Bouyahiaoui R, Nazih R et al (2018) Characterization of the onset of puberty in Tazegzawt lambs, an endangered Algerian sheep: body weight, thoracic perimeter, testicul-ar growth, and seminal parameters. Vet World 11(7):889–894. https://doi.org/10.14202/vetworld.2018.889-894
Patterson JL, Beltranena E, Foxcroft GR (2010) The effect of gilt age at first estrus and br-eeding on third estrus on sow body weight changes and long-term reproductive performance. J Anim Sci 88(7):2500–2513. https://doi.org/10.2527/jas.2008-1756
Yang GR, Chen LH, Grant GR et al (2016) Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med 8(324):324. https://doi.org/10.1126/scitranslmed.aad3305
Li ZJ, Li YS, Ren Y, Li CM (2020) High ambient temperature disrupted the circadian rhyt-hm of reproductive hormones and changed the testicular expression of steroidogenesis genes and clock genes in male mice. Mol Cell Endocrinol 500(13):110639. https://doi.org/10.1016/j.mce.2019.110639
Meyer V (2014) Searching for the testicular clock: comparing daily patterns of clock gene expression and the duration of the seminiferous epithelium cycle in three hamster species. Dissertation, SES School of Engineering and Science
Shimizu T, Hirai Y, Murayama C, Miyamoto A, Miyazaki H, Miyazaki K (2011) Circadian Clock genes Per2 and clock regulate steroid production, cell proliferation, and luteinizing hormone receptor transcription in ovarian granulosa cells. Biochem Biophys Res Commun 412(1):132–135. https://doi.org/10.1016/j.bbrc.2011.07.058
Baburski AZ, Andric SA, Kostic TS (2019) Luteinizing hormone signaling is involved in synchronization of Leydig cell’s clock and is crucial for rhythm robustness of testosterone pro-duction†. Biol Reprod 100(5):1406–1415. https://doi.org/10.1093/biolre/ioz020
Mereness AL, Murphy ZC, Forrestel AC, Susan B, Ko C, Richards JS, Sellix MT (2016) Conditional deletion of Bmal1 in ovarian theca cells disrupts ovulation in female mice. Endocrinology 2:913–927. https://doi.org/10.1210/en.2015-1645
Zhang JO, Liu JS, Zhu K et al (2016) Effects of BMAL1–SIRT1-positive cycle on estrogen synthesis in human ovarian granulosa cells: an implicative role of BMAL1 in PCOS. Endocrine 53(2):574–584. https://doi.org/10.1007/s12020-016-0961-2
Li RW, Cheng ST, Wang ZR (2015) Circadian clock gene plays a key role on ovarian cycle and spontaneous abortion. Cell Physiol Biochem 37(3):911–920. https://doi.org/10.1159/000430218
Cheng ST, Liang X, Wang YH et al (2016) The circadian Clock gene regulates acrosin acti-vity of sperm through serine protease inhibitor A3K. Exp Biol Med 241(2):205. https://doi.org/10.1177/1535370215597199
Yang JB, Zhang ZW, Zhang YY et al (2018) CLOCK interacts with RANBP9 and is invol-ved in alternative splicing in spermatogenesis. Gene 642:199–204. https://doi.org/10.1016/j.gene.2017.11.007
Li C, Xiao SW, Hao J, Liao XG, Li G (2018) Cry1 deficiency leads to testicular dysfuncti-on and altered expression of genes involved in cell communication, chromatin reorganization, spermatogenesis, and immune response in mouse testis. Mol Reprod Dev 85(4):325–335. https://doi.org/10.1002/mrd.22968
Dimova EY, Jakupovic M, Kubaichuk K et al (2019) The circadian clock protein CRY1 is a negative regulator of HIF-1α. iScience 13:284–304. https://doi.org/10.1016/j.isci.2019.02.027
Zhao SQ, Gao Y, Zhang Y, Yang XP, Yang Z (2021) cAMP/PKA/CREB signaling pathway-mediated effects of melatonin receptor genes on clock gene expression in bactrian camel ovarian granulosa cells. Domest Anim Endocrinol 76(4):106609. https://doi.org/10.1016/j.domaniend.2021.106609
Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32(suppl_1):D91–D94. https://doi.org/10.1093/nar/gkh012
Chen CJ, Xia R, Chen H et al (2020) TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Bo DD, Jiang XP, Liu GQ et al (2020) Multipathway synergy promotes testicular transition from growth to spermatogenesis in early-puberty goats. BMC Genomics 21(1):372. https://doi.org/10.1186/s12864-020-6767-x
Wang SF, Zhao SQ, Liu CP et al (2021) Expression and location of Cry1 gene in reprod-uctive axis of male sheep. Chin J Anim Sci 57(03):97–103. https://doi.org/10.19556/j.0258-7033.20200506-04
Jordan SD, Kriebs A, Vaughan M et al (2017) CRY1/2 selectively repress PPARδ and limit exercise capacity. Cell Metab 26(1):243–255. https://doi.org/10.1016/j.cmet.2017.06.002
Regueira M, Riera MF, Galardo MN et al (2015) FSH and bFGF regulate the expression of genes involved in Sertoli cell energetic metabolism. Gen Comp Endocrinol 222(2766):124–133. https://doi.org/10.1016/j.ygcen.2015.08.011
Orth JM, Gunsalus GL, Lamperti AA (1988) Evidence from sertoli cell-depleted rats indicates that spermatid number in adults depends on numbers of sertoli cells produced during perinatal development. Endocrinology 122(3):787–794. https://doi.org/10.1210/endo-122-3-787
Ni FD, Hao SL, Yang WX (2019) Multiple signaling pathways in Sertoli cells: recent findi-ngs in spermatogenesis. Cell Death Dis 10(8):541. https://doi.org/10.1038/s41419-019-1782-z
Zhou L, He J, Sun S, Yu YM, Zhang TQ, Wang MH (2019) Cryptochrome-1 regulates Os-teoblast differentiation via the AKT kinase and extracellular signal-regulated kinase signaling pathways. Cell Reprogram 21(3):141–151. https://doi.org/10.1089/cell.2018.0054
Chen JN (2019) Study on the function of CRY1 gene in yak. Dissertation, Gansu Agricultural University.
Xu K, Wang J, Liu HY, Zhao J, Lu WF (2020) Melatonin promotes the proliferation of chicken sertoli cells by activating the ERK/Inhibin alpha subunit signaling pathway. Molecules 25(5):1230. https://doi.org/10.3390/molecules25051230
Niu BW, Li B, Wu CY et al (2016) Melatonin promotes goat spermatogonia stem cells (SSCs) proliferation by stimulating glial cell line-derived neurotrophic factor (GDNF) production in Sertoli cells. Oncotarget 7(47):77532–77542. https://doi.org/10.18632/oncotarget.12720
Horst GTJVD, Muijtjens M, Kobayashi K et al (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398(6728):627–630. https://doi.org/10.1038/19323
Acknowledgements
We are thankful to Chenhui Liu, Dandan Du and all members of the Jiang laboratory who provided expertise that greatly assisted the research.
Funding
Supported by China Agriculture Research System of MOF and MARA (CARS-38). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization, XJ, and GL; Methodology, YH, YY and CL; Investigation, YH; Supervision, XJ; Writing—Original Draft, YH and XJ; Writing—Review & Editing, all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The main research project researcher of this work was approved for animal ethics by the Scientific Ethic Committee of Laboratory Animal Centre, Huazhong Agricultural University (HZAUGO-2018–006, 1 March 2018), China. All applicable guidelines for the care and use of animals were followed by the authors.
Consent for publication
The manuscript has been read and approved by all named authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, Y., Jiang, X., Yan, Y. et al. Expression of cell proliferation regulatory factors bricd5, tnfrsf21, cdk1 correlates with expression of clock gene cry1 in testes of Hu rams during puberty. Mol Biol Rep 48, 7379–7385 (2021). https://doi.org/10.1007/s11033-021-06747-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06747-6