Abstract
Background
Ashwagandha (Withania somnifera (L.) Dunal), popularly known as Indian ginseng or winter cherry is a multipurpose plant of immense therapeutic value in the ayurvedic and indigenous medicine system and distributed in wide geographic locations and exhibiting extensive phenotypic and chemical variability.
Methods and results
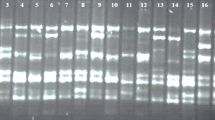
The present study was carried out to assess the molecular genetic diversity among 4 CIMAP varieties and five local cultivars of ashwagandha and cluster dendrograms were created by using 20 ISSR primers. A total of 224 bands of varied length were produced, out of which 193 (86.1%) products were polymorphic and 31 (13.8%) products were monomorphic. Where each ISSR arbitrary primer had 5–16 valuable bands with an average of 11.2 bands per primer, of which 86.16% bands were polymorphic. The PIC values ranged from 0.16 to 0.36 with an average PIC value of 0.29 and RP values ranged from 2.22 to 7.99. The UPGMA cluster analysis of 20 ISSR primers grouped the nine accessions into 2 major clusters. The first and second major cluster consists of seven and two accessions respectively.
Conclusion
Therefore, this study provides evidence that ISSR based molecular diversity assessment can be used as an efficient tool for detecting similarity and phylogenetic relationships among genotypes of Withania somnifera collected from different geographical locations. This information can be used to improve root and other characteristics of ashwagandha genotypes and there is also scope for the development of high-yielding varieties by selecting diverse parents for crossing (based on the molecular diversity) from the present accessions.


Similar content being viewed by others
Data availability
Not applicable.
Code availability
Not applicable.
Abbreviations
- AFLP:
-
Amplified Fragment Length Polymorphism
- CTAB:
-
Cetyl trimethyl Ammonium Bromide
- ISSR:
-
Inter Simple Sequence Repeat
- PCR:
-
Polymerase Chain Reaction
- PIC:
-
Polymorphism Information Content
- PPB:
-
Polymorphic Bands
- Rp:
-
Resolving Power
- SSR:
-
Simple Sequence Repeat
- UPGMA:
-
Unweighted Pair Group Method with Arithmetic mean
References
Chopra RN (1994) Glossary of Indian Medicinal Plants. Academic Publishers India, New Delhi
Devi PU, Sharada AC, Solomon FE (1993) Antitumor and radio sensitizing effects of Withania somnifera (Ashwagandha) on a transplantable mouse tumor, Sarcoma-180. Indian J Exp Biol. 31:607–11
Singh S, Kumar S (1998) Withania somnifera: The Indian ginseng Ashwagandha. Central Institute of Medicinal and Aromatic Plants, Lucknow (India Stony Brook, New York, NY.)
Srivastava A, Anil KG, Shanker K, Madan MG, Mishra R, Raj KL (2018) Genetic variability, associations, and path analysis of chemical and morphological traits in Indian ginseng [Withania somnifera (L.) Dunal] for selection of higher yielding genotypes. J Ginseng Res 42:158–164. https://doi.org/10.1016/j.jgr.2017.01.014
Gupta AK, Verma SR, Gupta MM, Saikia D, Verma RK, Jhang T (2011) Genetic diversity in germplasms collections of Withania somnifera for root and leaf alkaloids. J Trop Med Plants 12:59–69
Al-Anbari AK, Al-Zubadiy MW, Malik Dawood W (2015) Genetic diversity of some taxa of Cucurbitaceae family based on RAPD markers. Adv Life Sci Technol 73:7–11
Yang HB, Kang WH, Nahm SH, Kang BC (2015) Methods for Developing Molecular Markers. In: Koh Hee-Jong, Kwon Suk-Yoon, Thomson Michael (eds) Current Technologies in Plant Molecular Breeding. Springer Science+Business Media, Dordrecht, pp 15–50
Barvaliya P, Singh NK, Dharajiya D, Pachchigar K (2018) Elucidation of genetic diversity among ashwagandha [Withania somnifera (L.) Dunal] genotypes using EST-SSR markers. Res. J. Biotech. 13(10):52–59
Henareh MB, Abdollahi HK (2016) Assessment of genetic diversity in tomato landraces using ISSR markers. Genetika 48(1):25–35
Ebrahimi M, Farajpour M, Rahimmalek M (2012) Inter- and intra-specific genetic diversity of Iranian yarrow species Achillea santolina and Achillea tenuifolia based on ISSR and RAPD markers. Genet Mol Res. https://doi.org/10.4238/2012.August.27.1
Farajpour M, Ebrahimi M, Amiri R, Golzari R, Sanjari S (2012) Assessment of genetic diversity in Achillea millefolium accessions from Iran using ISSR marker. Biochem Syst Ecol 43:73–79. https://doi.org/10.1016/j.bse.2012.02.017
Maleki M, Shojaeiyan A, Monfared SR (2018) Population structure, morphological and genetic diversity within and among melon (Cucumis melo L.) landraces in Iran. J Genet Eng Biotechnol 16:599–606
Baruah J, Pandey SK, Begum T, Sarma N, Paw M, Lal M (2019) Molecular diversity assessed amongst high dry rhizome recovery ginger germplasm (Zingiber officinale roscoe) from NE-India using RAPD and ISSR markers. Ind Crop Prod 129:463–471. https://doi.org/10.1016/j.indcrop.2018.12.037
Saxena A (2014) Vol TR-RJ of B, 2020 U. assessment of genetic diversity in cowpea (Vigna unguiculata L. Walp.) germplasm. Bull Inst Trop Agric Kyushu Univ 37:57–63. https://doi.org/10.11189/bita.37.57
Ghaffar K, Mohammad S (2017) Genetic Diversity in Tomato Varieties Assessed by ISSR Markers. International Journal of Vegetable Science 24(4):353–360. https://doi.org/10.1080/19315260.2017.1419397
Farajpour M, Ebrahimi M, Amiri R, Noori SAS, Sanjari S, Golzari R (2011) Study of genetic variation in yarrow using inter-simple sequence repeat (ISSR) and random amplified polymorphic DNA (RAPD) markers. Afr J Biotech 10:73–86
Khanuja SPS, Shasany AK, Darokar MP, Kumar S (1999) Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Mol Biol Report 17(1):1–7
Roldan RJ, Dendauw E, Van B, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134
Prevost A, Wilkinson M (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107. https://doi.org/10.1007/s001220051046
Rohlf FJ (1992) NTSYS-pc: Numerical taxonomy and multivariate analysis system. ver. 1.70.
Pan L, Fu J, Zhang R et al (2017) Genetic diversity among germplasms of Pitaya based on SSR markers. Scientia Horticulturae 225:171–176
Wolt JD (2019) Current risk assessment approaches for environmental and food and feed safety assessment. Transgenic Res 28:111–117. https://doi.org/10.1007/s11248-019-00140-7
Bilal AM, Sushma K, Kumar Arun, Kaul MK, Sooda AS, Raina SN (2009) Assessment and characterization of genetic diversity in Withania somnifera (L.) Dunal using RAPD and AFLP markers. African Journal of Biotechnology 10(66):14746–14756
Patel HK, Fougat RS, Kumar S, Mistry JG, Kumar M (2015) Detection of genetic variation in Ocimum species using RAPD and ISSR markers. 3 Biotech 5:697–707
Gelotar MJ, Dharajiya DT, Solanki SD, Prajapati NN, Kapil KT (2019) Genetic diversity analysis and molecular characterization of grain amaranth genotypes using inter simple sequence repeat (ISSR) markers. Bull Natl Res Cent. https://doi.org/10.1186/s42269-019-0146-2
Liu X, Jun D, Asaduzzaman Khan MD, Jingliang C, Chunli W, Zhiqiang M, Hanchun C, Tao H, Junjiang F (2020) Analysis of genetic diversity and similarities between different Lycium varieties based on ISSR analysis and RAMP-PCR markers. World Academy of Sci J 2:83–90
Gilberto CL, Danuza KS, Claudete AM, Maria de Fátima PSM (2020) ISSR markers to assess genetic diversity of cultivated populations from artificial selection of Stevia rebaudiana (Bert.) Bertoni. Breed Sci 70:508–514. https://doi.org/10.1270/jsbbs
Khodaee L, Azizinezhad R, Etminan AR, Khosroshahi Mahmoud (2021) Assessment of genetic diversity among Iranian Aegilops triuncialis accessions using ISSR, SCoT, and CBDP markers. J Genet Eng Biotechnol. https://doi.org/10.1186/s43141-020-00107-w
Niraj T, Navinder S, Vandana M, Sunil Kumar ST (2012) Assessment of genetic diversity among Withania somnifera collected from central India using RAPD and ISSR analysis. Medicinal and aromatic plant science and biotechnology 6(1):33–39
Žiarovská J, Ražná V, Labajová M (2013) Using of inter microsatellite polymorphism to evaluate gamma-irradiated Amaranth mutants. Emir J Food Agric 25(9):673–681
Pfeiffer T, Roschanski AM, Pannell Korbecka G, Schnitter M (2011) Characterization of microsatellite loci and reliable genotyping in a polyploidy plant, Mercurialis perennis (Euphorbiaceae). J. Hered. 102:479–488
Dharmar K, De John B (2011) RAPD analysis in genetic variability in wild populations of Withania somnifera (L.) Dunal. International journal of biological technology 2:21–25
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Tiwari SK, Karihaloo JL, Hameed N, Ambika BG (2009) Molecular Characterization of Brinjal (Solanum melongena L) Cultivars using RAPD and ISSR Markers. J Plant Biochem Biotechnol 18:189–195. https://doi.org/10.1007/BF03263318
Shahana K, Rehana AS (2016) Assessment of genetic diversity among India Ginseng, Withania somnifera (L) Dunal using RAPD and ISSR markers. Research in Biotechnology 7:01–10
Nayak S, Naik PK, Acharya L, Mukherjee AK, Panda PC, Das P (2005) Assessment of genetic diversity among 16 promising cultivars of ginger using cytological and molecular markers. Z Natur for schung 60:485–492
Kanika B, Sunita K, Anita P, Om Prakash Y (2013) Genetic Variability Analysis Using ISSR Markers in Withania Somnifera L. Dunal Genotypes from Different Regions. J Herbs Spices Med Plants 19(1):22–32. https://doi.org/10.1080/10496475.2012.734770
Acknowledgements
The authors acknowledge the Director CSIR-CIMAP for providing the facility for conducting research work.
Funding
No Founding sources.
Author information
Authors and Affiliations
Contributions
CH conceived the research idea and designed the experiments, and performed, analyzed the data, and wrote the manuscript. RP performed the experiments VS gave technical guidance while writing the manuscript and gave lab consumables for doing work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
CIMAP Publication No. CIMAP/PUB/2020/FEB /09.
Rights and permissions
About this article
Cite this article
Hiremath, C., Philip, R. & Sundaresan, V. Molecular characterization of India Ginseng Withania somnifera (L) using ISSR markers. Mol Biol Rep 48, 3971–3977 (2021). https://doi.org/10.1007/s11033-021-06397-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06397-8