Abstract
Background
Teleost scale not only provides a protective layer resisting penetration and pathogens but also participate in coloration. It is interesting to study the mechanism of teleost scale formation. Furthermore, whether there existed consensus genes between scale coloration and skin coloration has not been examined yet.
Methods and results
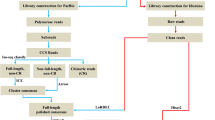
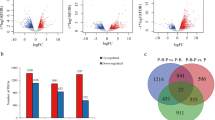
We analyzed the transcriptome profiles of red scale, white scale, red skin, and white skin of common carp (Cyprinus carpio). Pair-wise comparison identified 3391 differentially expressed genes (DEGs) between scale and skin, respectively. The 1765 up-regulated genes (UEGs) in scale, as the down-regulated genes in skin, preferred mineralization and other scale development-related processes. The 1626 skin UEGs were enriched in the morphogenesis of skin and appendages. We also identified 195 UEGs in white scale and 223 UEGs in red scale. The white scale UEGs primarily participated in regulation of growth and cell migration. The UEGs in red scale preferred pigment cell differentiation and retinoid metabolic process. A total of 22 DEGs had consensus expression patterns in skin and scale of the same coloration. The expression levels of these DEGs clearly grouped skin and scale of the same coloration together with principle component analysis and correlation analysis. Eleven consensus DEGs were homologous to the orthologs of Poropuntius huangchuchieni, 82% of which were under strong purifying selection. Eight processes including lipid storage and lipid catabolism were shared in both scale pigmentation and skin pigmentation.
Conclusions
We identified consensus DEGs and biological processes in scale and skin pigmentation. Our transcriptome analysis will contribute to further elucidation of mechanisms of teleost scale formation and coloration.



Similar content being viewed by others
Data availability
The raw RNA sequencing data were deposited in NCBI SRA database (BioProject: PRJEB19194). The updated common carp genes were deposited at DDBJ/EMBL/GenBank under the accession of GFWU00000000. The protein sequences, GO terms, and gtf file were available at figshare (https://figshare.com/s/d34e4ad1ba113c9b4b09, https://figshare.com/s/150a49fc5f62522199b7, and https://figshare.com/s/e5faf21db6bfbba16a34).
References
Zhu D, Ortega CF, Motamedi R, Szewciw L, Vernerey F, Barthelat F (2012) Structure and mechanical performance of a “modern” fish scale. Adv Eng Mater 14(4):B185–B194. https://doi.org/10.1002/adem.201180057
Sharpe PT (2001) Fish scale development: hair today, teeth and scales yesterday? Curr Biol 11(18):R751–R752. https://doi.org/10.1016/S0960-9822(01)00438-9
Schonborner AA, Boivin G, Baud CA (1979) The mineralization processes in teleost fish scales. Cell Tissue Res 202(2):203–212
Nusslein-Volhard C, Singh AP (2017) How fish color their skin: a paradigm for development and evolution of adult patterns: multipotency, plasticity, and cell competition regulate proliferation and spreading of pigment cells in Zebrafish coloration. BioEssays. https://doi.org/10.1002/bies.201600231
Braasch I, Schartl M, Volff JN (2007) Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol Biol 7:74. https://doi.org/10.1186/1471-2148-7-74
Haffter P, Odenthal J, Mullins MC, Lin S, Farrell MJ, Vogelsang E, Haas F, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Hopkins N, Nüsslein-Volhard C (1996) Mutations affecting pigmentation and shape of the adult zebrafish. Dev Genes Evol 206(4):260–276. https://doi.org/10.1007/s004270050051
Bar I, Kaddar E, Velan A, David L (2013) Melanocortin receptor 1 and black pigmentation in the Japanese ornamental carp (Cyprinus carpio var. Koi). Front Genet 4:6. https://doi.org/10.3389/fgene.2013.00006
Li X-M, Song Y-N, Xiao G-B, Zhu B-H, Xu G-C, Sun M-Y, Xiao J, Mahboob S, Al-Ghanim KA, Sun X-W, Li J-T (2015) Gene expression variations of red-white skin coloration in common carp (Cyprinus carpio). Int J Mol Sci 16(9):21310–21329. https://doi.org/10.3390/ijms160921310
Neave F (1940) Memoirs: on the histology and regeneration of the teleost scale. Q J Microsc Sci 2–81(324):541
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622. https://doi.org/10.1373/clinchem.2008.112797
Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11(1):485. https://doi.org/10.1186/1471-2105-11-485
Xu P, Zhang X, Wang X, Li J, Liu G, Kuang Y, Xu J, Zheng X, Ren L, Wang G, Zhang Y, Huo L, Zhao Z, Cao D, Lu C, Li C, Zhou Y, Liu Z, Fan Z, Shan G, Li X, Wu S, Song L, Hou G, Jiang Y, Jeney Z, Yu D, Wang L, Shao C, Song L, Sun J, Ji P, Wang J, Li Q, Xu L, Sun F, Feng J, Wang C, Wang S, Wang B, Li Y, Zhu Y, Xue W, Zhao L, Wang J, Gu Y, Lv W, Wu K, Xiao J, Wu J, Zhang Z, Yu J, Sun X (2014) Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet 46(11):1212–1219. https://doi.org/10.1038/ng.3098
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360. https://doi.org/10.1038/nmeth.3317
Wang L, Wang S, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28(16):2184–2185. https://doi.org/10.1093/bioinformatics/bts356
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295. https://doi.org/10.1038/nbt.3122
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8(8):1494–1512. https://doi.org/10.1038/nprot.2013.084
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578. https://doi.org/10.1038/nprot.2012.016
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106. https://doi.org/10.1186/gb-2010-11-10-r106
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Chen C, Xia R, Chen H, He Y (2018) TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. https://doi.org/10.1101/289660
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607-d613. https://doi.org/10.1093/nar/gky1131
Savan R, Igawa D, Sakai M (2003) Cloning, characterization and expression analysis of interleukin-10 from the common carp, Cyprinus carpio L. Eur J Biochem 270(23):4647–4654. https://doi.org/10.1046/j.1432-1033.2003.03854.x
Wang X, Gan X, Li J, Chen Y, He S (2016) Cyprininae phylogeny revealed independent origins of the Tibetan Plateau endemic polyploid cyprinids and their diversifications related to the Neogene uplift of the plateau. Sci China Life Sci 59(11):1149–1165. https://doi.org/10.1007/s11427-016-0007-7
Barron MJ, Brookes SJ, Draper CE, Garrod D, Kirkham J, Shore RC, Dixon MJ (2008) The cell adhesion molecule nectin-1 is critical for normal enamel formation in mice. Hum Mol Genet 17(22):3509–3520. https://doi.org/10.1093/hmg/ddn243
Brancati F, Fortugno P, Bottillo I, Lopez M, Josselin E, Boudghene-Stambouli O, Agolini E, Bernardini L, Bellacchio E, Iannicelli M, Rossi A, Dib-Lachachi A, Stuppia L, Palka G, Mundlos S, Stricker S, Kornak U, Zambruno G, Dallapiccola B (2010) Mutations in PVRL4, encoding cell adhesion molecule nectin-4, cause ectodermal dysplasia-syndactyly syndrome. Am J Hum Genet 87(2):265–273. https://doi.org/10.1016/j.ajhg.2010.07.003
Whyte MP, McAlister WH, Fallon MD, Pierpont ME, Bijanki VN, Duan S, Otaify GA, Sly WS, Mumm S (2017) Raine syndrome (OMIM #259775), caused by FAM20C mutation, is congenital sclerosing osteomalacia with cerebral calcification (OMIM 259660). J Bone Miner Res 32(4):757–769. https://doi.org/10.1002/jbmr.3034
Vogel P, Hansen GM, Read RW, Vance RB, Thiel M, Liu J, Wronski TJ, Smith DD, Jeter-Jones S, Brommage R (2012) Amelogenesis imperfecta and other biomineralization defects in Fam20a and Fam20c null mice. Vet Pathol 49(6):998–1017. https://doi.org/10.1177/0300985812453177
Bolanos A, Hotton D, Ferbus D, Loiodice S, Berdal A, Babajko S (2012) Regulation of calbindin-D(28k) expression by Msx2 in the dental epithelium. J Histochem Cytochem 60(8):603–610. https://doi.org/10.1369/0022155412450641
Arduini BL, Gallagher GR, Henion PD (2008) Zebrafish endzone regulates neural crest-derived chromatophore differentiation and morphology. PLoS ONE 3(7):e2845. https://doi.org/10.1371/journal.pone.0002845
Sefc KM, Brown AC, Clotfelter ED (2014) Carotenoid-based coloration in cichlid fishes. Comp Biochem Physiol A 173:42–51. https://doi.org/10.1016/j.cbpa.2014.03.006
Nikčević G, Savić T, Kovačević-Grujičić N, Stevanović M (2008) Up-regulation of the SOX3 gene expression by retinoic acid: characterization of the novel promoter-response element and the retinoid receptors involved. J Neurochem 107(5):1206–1215. https://doi.org/10.1111/j.1471-4159.2008.05670.x
Suzuki R, Miyahara K, Murakami H, Doi T, Lane GJ, Mabuchi Y, Suzuki N, Yamataka A, Akazawa C (2014) Abnormal neural crest innervation in Sox10-Venus mice with all-trans retinoic acid-induced anorectal malformations. Pediatr Surg Int 30(2):189–195. https://doi.org/10.1007/s00383-013-3452-z
Bastonini E, Kovacs D, Picardo M (2016) Skin pigmentation and pigmentary disorders: focus on epidermal/dermal cross-talk. Ann Dermatol 28(3):279–289. https://doi.org/10.5021/ad.2016.28.3.279
Kelsh RN (2004) Genetics and evolution of pigment patterns in fish. Pigment Cell Res 17(4):326–336. https://doi.org/10.1111/j.1600-0749.2004.00174.x
Patel S, Huang Y-W, Reheman A, Pluthero Fred G, Chaturvedi S, Mukovozov Ilya M, Tole S, Liu G-Y, Li L, Durocher Y, Ni H, Kahr Walter HA, Robinson Lisa A (2012) The cell motility modulator Slit2 is a potent inhibitor of platelet function. Circulation 126(11):1385–1395. https://doi.org/10.1161/CIRCULATIONAHA.112.105452
Tanno T, Fujiwara A, Sakaguchi K, Tanaka K, Takenaka S, Tsuyama S (2007) Slit3 regulates cell motility through Rac/Cdc42 activation in lipopolysaccharide-stimulated macrophages. FEBS Lett 581(5):1022–1026. https://doi.org/10.1016/j.febslet.2007.02.001
Meyer-Rochow VB, Royuela M, Fraile B, Paniagua R (2001) Smooth muscle proteins as intracellular components of the chromatophores of the Antarctic fishes Pagothenia borchgrevinki and Trematomus bernacchii (Nototheniidae). Protoplasma 218(1–2):24–30
McIntyre LM, Lopiano KK, Morse AM, Amin V, Oberg AL, Young LJ, Nuzhdin SV (2011) RNA-seq: technical variability and sampling. BMC Genomics 12:293
Mundy Nicholas I, Stapley J, Bennison C, Tucker R, Twyman H, Kim K-W, Burke T, Birkhead Tim R, Andersson S, Slate J (2016) Red carotenoid coloration in the zebra finch is controlled by a cytochrome P450 gene cluster. Curr Biol 26(11):1435–1440. https://doi.org/10.1016/j.cub.2016.04.047
Lee SA, Jiang H, Trent CM, Yuen JJ, Narayanasamy S, Curley RW Jr, Harrison EH, Goldberg IJ, Maurer MS, Blaner WS (2014) Cardiac dysfunction in beta-carotene-15,15′-dioxygenase-deficient mice is associated with altered retinoid and lipid metabolism. Am J Physiol Heart Circ Physiol 307(11):H1675-1684. https://doi.org/10.1152/ajpheart.00548.2014
Shiraki T, Kojima D, Fukada Y (2010) Light-induced body color change in developing zebrafish. Photochem Photobiol Sci 9(11):1498–1504. https://doi.org/10.1039/c0pp00199f
Ma W, Zhu ZH, Bi XY, Murphy RW, Wang SY, Gao Y, Xiao H, Zhang YP, Luo J (2014) Allopolyploidization is not so simple: evidence from the origin of the tribe Cyprinini (Teleostei: Cypriniformes). Curr Mol Med 14(10):1331–1338. https://doi.org/10.2174/1566524014666141203101543
Zhang Z, Li J, Zhao X-Q, Wang J, Wong GK-S, Yu J (2006) KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4(4):259–263. https://doi.org/10.1016/S1672-0229(07)60007-2
Jeong H, Lim KM, Goldenring JR, Nam KT (2019) Rab25 deficiency perturbs epidermal differentiation and skin barrier function in mice. Biomol Ther 27(6):553–561. https://doi.org/10.4062/biomolther.2019.125
Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J, Oppenheim JJ (2003) Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J Leukoc Biol 74(3):448–455. https://doi.org/10.1189/jlb.0103024
Denis C, Deiteren K, Mortier A, Tounsi A, Fransen E, Proost P, Renauld J-C, Lambeir A-M (2012) C-Terminal clipping of chemokine CCL1/I-309 enhances CCR8-mediated intracellular calcium release and anti-apoptotic activity. PLoS ONE 7(3):e34199. https://doi.org/10.1371/journal.pone.0034199
Hromas R, Broxmeyer HE, Kim C, Nakshatri H, Christopherson K 2nd, Azam M, Hou YH (1999) Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem Biophys Res Commun 255(3):703–706. https://doi.org/10.1006/bbrc.1999.0257
Borel P (2003) Factors affecting intestinal absorption of highly lipophilic food microconstituents (fat-soluble vitamins, carotenoids and phytosterols). Clin Chem Lab Med 41(8):979–994. https://doi.org/10.1515/cclm.2003.151
Acknowledgements
This study is supported by National Natural Science Foundation of China (31672644) and the Special Scientific Research Funds for Central Non-profit Institutes, Chinese Academy of Fishery Sciences (2020XT0103 and 2020TD24).
Author information
Authors and Affiliations
Contributions
JLT conceived and designed the study and all the experiments. YJZ and JX performed the experiments and collected all data. YJZ and JX analyzed the data. JLT and YJZ wrote the manuscript. YJZ, MDHY and RZ edited the figures and tables. QW and YZ participated in discussion. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
Use of all these samples was approved by the Institutional Animal Care and Use Committee of Key Laboratory of Aquatic Genomics, Chinese Academy of Fishery Science (Animal Ethics No. IACUC-201501).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, YJ., Xiao, J., Huangyang, MD. et al. Transcriptome sequencing and analysis for the pigmentation of scale and skin in common carp (Cyprinus carpio). Mol Biol Rep 48, 2399–2410 (2021). https://doi.org/10.1007/s11033-021-06273-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06273-5