Abstract
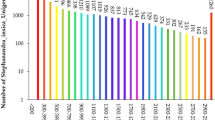
High-throughput sequencing of the Phoebe bournei transcriptome was performed, and novel SSR markers were identified. A total of 73,518 nonredundant unigenes were assembled and annotated by sequence similarity searching in diverse public databases. A total of 40,853 SSRs were identified from 73,518 unigenes. Twenty-three pairs of polymorphic EST-SSR markers were selected from 98 markers and used for genetic analyses in 75 individuals from three P. bournei populations. The 23 pairs of markers could detect abundant genetic information from the samples (PIC = 0.769), and cross-species amplification was successfully performed in other related species. Three populations had high level of genetic diversity (He = 0.658 in average), of which the population YS from Jiangxi province had the most abundant genetic diversity (He = 0.722). The results of genetic structure analyses showed that the population YS from Jiangxi province had obvious genetic differences from the other two populations, and the genetic information of the population SX from Fujian province was related to that of the population LC from Guangdong province and the population YS. The transcriptomic resources and EST-SSR markers are valuable tools not only for the ecological conservation of P. bournei but also for phylogenetic studies.



Similar content being viewed by others
References
Chen Z (2012) Development and application of microsatellite markers in Betula luminifera. Dissertation, Zhejiang Agriculture and Forestry University
Chen SP, Sun WH, Xiong YF, Jiang YT, Liu XD, Liao XY, Zhang DY, Jiang SZ, Li Y, Liu B, Ma L, Yu X, He L, Liu B, Feng JL, Feng LZ, Wang ZW, Zou SQ, Lan SR, Liu ZJ (2020) The Phoebe genome sheds light on the evolution of magnoliids. Hortic Res 7:146
Cheng XM, Huang XX (2011) Development and application of SSR markers in plants. Chin Agr Sci Bull 27(5):304–307
Ding YJ, Zhang JH, Lu YF, Lin EP, Lou LH, Tong ZK (2015) Development of EST-SSR markers and analysis of genetic diversity in natural populations of endemic and endangered plant Phoebe chekiangensis. Biochem Syst Ecol 63(63):183–189
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Ge YJ, Liu YJ, Shen AH, Lin XC (2015) Fengshui forests conserve genetic diversity: a case study of Phoebe bournei (Hemsl.) Yang in southern China. Genet Mol Res 14(1):1986
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652
He YH, Liang RL, Jiang Y, Sun B (2013) Research progress and development countermeasures of rare tree species Phoebe bournei. Guangxi For Sci 42(04):365–370
Huang YQ, Yin GT, Yang JC, Yu N, Zou WT, Li RS (2020) Construction of core collection of Phoebe bournei based on SSR molecular markers. Mol Plant Breed 18(08):2641–2648
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806
Jiang XM, Wen Q, Ye JS, Xiao FM, Jiang M (2009) RAPD analysis of genetic diversity of Phoebe bournei natural population. Acta Ecol Sin 29(1):438–444
Jin JQ, Cui HR, Chen WY, Lu MZ, Yao YL, Xin Y, Gong XC (2006) Data mining for SSRs in ESTs and development of EST-SSR marker in tea plant (Camellia sinensis). J Tea Sci 46(1):35–54
Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sala F, Van de Wiel C, Bredemeijer G, Vosman B, Matthes M, Daly A, Brettschneider R, Bettini P, Buiatti M, Maestri E, Malcevschi A, Marmiroli N, Aert R, Volckaert G, Rueda J, Linacero R, Vazquez A, Karp A (1997) Reproducibility of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed 3(5):381–390
Li ZH, Li BH, Qi CJ, Yu XL, Wu Y (2012) Studies on importance of valuable wood species resources and its development strategy. J Cent S Univ For Technol 32(11):1–8
Li YG, Xu WQ, Zou WT, Jiang DY, Liu XH (2017) Complete chloroplast genome sequences of two endangered Phoebe (Lauraceae) species. Bot Stud 58:37
Li J, Dong LJ, Lin JY, Liu XS, Liang RL (2019) ISSR analysis of genetic diversity of Phoebe bournei from different geographical populations. Mol Plant Breed 17(23):7822–7828
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Liu YX, Song XC, Jiang XM (2013) Chromosome karyotype analysis of Phoebe neurantha and Phoebe bournei. J Nanjing For Univ 37(05):157–160
Liu CC, Dou Y, Guan XL, Fu Q, Zhang Z, Hu ZH, Zheng J, Lu YZ, Li W (2017) De novo transcriptomic analysis and development of EST-SSRs for Sorbus pohuashanensis (Hance) Hedl. PLoS ONE 12(6):e0179219
Liu D, Liu B, Zeng QM, Chen SP, Liu B, Li Y (2019) SSR analysis of the genetic difference of Phoebe Phoebe superior genotypes. J For Environ 39(05):449–453
Lu YF, Yang AN, Zhang JH, Lou LH, Huang HH, Tong ZK (2018) Development and transferability evaluation of EST-SSR markers based on transcriptome data of Phoebe sheareri. J Agric Biotechnol 26(6):1014–1024
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol 4(1):137–138
Shi XD, Zhu XH, Sheng YZ, Zhuang GQ, Chen F (2016) Development of SSR markers based on transcriptome sequence of Phoebe zhennan. Sci Silvae Sin 52(11):71–78
Wang D, Cao LY, Gao JP (2014) Data mining of simple sequence repeats in Codonopsis pilosula transcriptome. Chin Tradit Herb Drugs 45(16):2390–2394
Wu SH, Jin H, Lai JX (2016) Genetic diversity of Fraxinus chinensisusing SSR makers. Inner Mongolia For Sci Technol 42(2):26–29
Xu M, Li HG (2008) Development and characterization of microsatellite markers from expressed sequence tags for Liriodendron. Mol Plant Breed 6(3):615–618
Xu XL, Xu LA, Huang MR, Wang ZR (2004) Genetic diversity of microsatellites (SSRs) of natural populations of Quercus variabilis. Hereditas 26(5):683–688
Yi M, Zhang L, Lei L, Cheng ZS, Sun SW, Lai M (2020) SSR analysis of Pinus elliottii transcriptome and development of EST-SSR marker. J Nanjing For Univ 44(2):75–83
Zerbo G, Cleophas Konrad H, Ouedraogo M, Geburek T (2017) Fourteen simple-sequence repeats newly developed for population genetic studies in Prosopis africana (Fabaceae Mimosoideae). BMC Res notes 10(1):1–5
Zhang R, Zhou ZC, Jin GQ, Wang SH, Wang XH (2012) Genetic diversity and differentiation within three species of the family Lauraceae in southeast China. Biochem Syst Ecol 44:317
Zhou Q, Mu KM, Ni ZX, Liu XH, Li YG, Xu LA (2020) Analysis of genetic diversity of ancient Ginkgo populations using SSR markers. Ind Crop Prod 145:111942
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR) anchored polymerase chain reaction amplification. Genomics 20(2):176–183
Acknowledgements
This study was supported by the National Key Research & Development Program (No. 2016YFD0600603) of China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study does not require ethical statement.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Q., Zhou, PY., Zou, WT. et al. EST-SSR marker development based on transcriptome sequencing and genetic analyses of Phoebe bournei (Lauraceae). Mol Biol Rep 48, 2201–2208 (2021). https://doi.org/10.1007/s11033-021-06228-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06228-w