Abstract
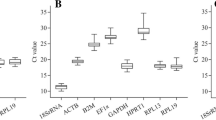
Tropomyosin, a muscle tissue protein is a major allergen in most of shellfish including mud crab. Quantitative real time-PCR (qRT-PCR) using a stable reference gene is the most sensitive approach to produce accurate relative gene expression that has yet to be demonstrated for allergenic tropomyosin in mud crab species. This study was conducted to identify the suitable reference gene and tropomyosin expression in different body parts of local mud crabs, Scylla olivacea, Scylla paramamosain and Scylla tranquebarica. Myosin, 18S rRNA, GADPH and EF1α were selected as candidate reference genes and their expression was measured in the abdomen, walking leg and cheliped tissues of local Scylla spp. The expression stability was analyzed using the comparative delta-Ct method, BestKeeper, NormFinder and geNorm then comprehensively ranked by RefFinder algorithm. Findings showed that EF1α was the most suitable reference gene across three mud crab species. Meanwhile, the abdomen, walking leg and cheliped selected their own suitable reference gene either Myosin, 18S rRNA, EF1α or GADPH. Overall, tropomyosin was the highest in S. tranquebarica, whereas the least was in S. paramamosain. Interestingly, tropomyosin was the highest in the abdomen of all mud crab species. This is the first analysis on reference genes selection for qRT-PCR data normalization of tropomyosin expression in mud crab. These results will provide more accurate findings for further gene expression and allergen analysis in Scylla spp.



Similar content being viewed by others
References
Guttman-Yassky E, Diaz A, Pavel AB et al (2019) Use of tape strips to detect immune and barrier abnormalities in the skin of children with early-onset atopic dermatitis. JAMA Dermatol 155(12):1358–1370. https://doi.org/10.1001/jamadermatol.2019.2983
Wang L, Zhang X, Zhang J et al (2018) Proteomic analysis and identification of possible allergenic proteins in mature pollen of Populus tomentosa. Int J Mol Sci 19(1):2–15. https://doi.org/10.3390/ijms19010250
Kuang J, Yan X, Genders AJ et al (2018) An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS ONE 13(5):1–27. https://doi.org/10.1371/journal.pone.0196438
Bustin SA, Benes V, Garson JA et al (2009) The MIQE Guidelines : Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(2):611–622. https://doi.org/10.1373/clinchem.2008.112797
Shekhar MS, Kiruthika J, Ponniah AG (2013) Identification and expression analysis of differentially expressed genes from shrimp (Penaeus monodon) in response to low salinity stress. Fish Shellfish Immunol 35(6):1957–1968. https://doi.org/10.1016/j.fsi.2013.09.038
Han K, Dai Y, Zhang Z et al (2018) Molecular Characterization and expression profiles of Sp-uchl3 and Sp-uchl5 during gonad development of Scylla paramamosain. Molecules 23(213):1–15. https://doi.org/10.3390/molecules23010213
Park K, Jo H, Kim DK et al (2019) Environmental pollutants impair transcriptional regulation of the Vitellogenin gene in the burrowing mud crab (Macrophthalmus japonicus). App Sci (Switzerland) 9(7):2–14. https://doi.org/10.3390/app9071401
Jiang H, Qian Z, Lu W et al (2015) Identification and characterization of reference genes for normalizing expression data from red swamp crawfish Procambarus clarkii. Int J Mol Sci 16(9):21591–21605. https://doi.org/10.3390/ijms160921591
Huang S, Chen X, Wang J et al (2017) Selection of appropriate reference genes for qPCR in the Chinese mitten crab, Eriocheir sinensis (Decapoda, Varunidae). Crustaceana 90(3):275–296. https://doi.org/10.1163/15685403-00003651
Hu Y, Fu H, Qiao H et al (2018) Validation and evaluation of reference genes for quantitative real-time PCR in Macrobrachium Nipponense. Int J Mol Sci 19(8):2258–2274. https://doi.org/10.3390/ijms19082258
Azra MN, Ikhwanuddin M (2016) A review of maturation diets for mud crab genus Scylla broodstock: present research, problems and future perspective. Saudi J Biol Sci 23(2):257–267. https://doi.org/10.1016/j.sjbs.2015.03.011
Ain N, Sharif M, Amalia N et al (2019) Species diversity and distribution of mud crab in Marudu Bay mangrove forest reserve, Sabah. Malaysia Borneo J Marine Sci Aqu 3(1):18–24
Haidr MA, Rosmilah M, Som CS et al (2018) Bioaccumulation of heavy metals in orange mud crab (Scylla olivacea) from Sungai Merbok. Kedah Int J Res Pharm Sci 10(1):654–658
Naim DM, Nor SAM, Mahboob S (2019) Reassessment of species distribution and occurrence of mud crab (Scylla spp, Portunidae) in Malaysia through morphological and molecular identification. Saudi J Biol Sci 27(2):643–652. https://doi.org/10.1016/j.sjbs.2019.11.030
Muhd-Farouk H, Nurul HA, Abol-Munafi AB et al (2019) Development of ovarian maturations in orange mud crab, Scylla olivacea (Herbst, 1796) through induction of eyestalk ablation and methyl farnesoate. Arab J Basic App Sci 26(1):171–181. https://doi.org/10.1080/25765299.2019.1588197
Nurul Izzah A, Rosmilah M, Zailatul HMY (2015) Identification of major and minor allergens of mud crab (Scylla Serrata). Med Health 10(2):90–97
Misnan R, Abdul Rahman NI, Yadzir ZHM et al (2015) Characterization of major allergens of local mud crab (Scylla serrata). Sci Res J 12(1): 1–10. https://doi.org/https://doi.org/10.24191/srj.v12i1.5434
Misnan R, Murad S, Yadzir ZHM et al (2012) Identification of the major allergens of Charybdis feriatus (red crab) and its cross-reactivity with Portunus pelagicus (blue crab). Asian Pac J Allergy Immunol 30(4):285–293
Lopata AL, Kleine-Tebbe J, Kamath SD (2016) Allergens and molecular diagnostics of shellfish allergy: Part 22 of the Series Molecular Allergology. Allergo J 25(7):24–32. https://doi.org/10.1007/s4062901601242
Su X, Lu L, Li Y et al (2020) Reference gene selection for quantitative realtime PCR (qRT-PCR) expression analysis in Galium aparine L. PLoS ONE 15(2):1–12. https://doi.org/10.1371/journal.pone.0226668
Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20(7):1736–1737. https://doi.org/10.1105/tpc.108.061143
Keenan C, Davie P, Mann D (1998a) A revision of the genus Scylla De Haan, 1833 (Crustacea : Decapoda : Brachyura : Portunidae). Raffles Bull Zool 46(1):217–245
Ikhwanuddin M, Azmie G, Juariah HM et al (2011) Biological information and population features of mud crab, genus Scylla from mangrove areas of Sarawak. Malaysia Fish Res 108(2–3):299–306. https://doi.org/10.1016/j.fishres.2011.01.001
Knuckey A (1996) Maturity in male mud crabs, Scylla serrata and the use of mating scars as a functional indicator. J Crust Biol 16(3):487–495
Xie F, Xiao P, Chen D (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80(1):75–84. https://doi.org/10.1007/s11103-012-9885-2
Vandesompele J, Preter KD, Poppe B et al (2002) Accurate normalization of real-time quantitative RT -PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):1–12
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real- time quantitative PCR and the 2-ΔΔCT method. Methods 408:402–408. https://doi.org/10.1006/meth.2001.1262
Pfaffl MW, Tichopad A, Prgomet C et al (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 26(6):509–515
Berumen-Varela G, Palomino-Hermosillo YA, Bautista-Rosales PU et al (2020) Identification of reference genes for quantitative real-time PCR in different developmental stages and under refrigeration conditions in soursop fruits (Annona muricata L.). Sci Hortic 260:1–6. https://doi.org/10.1016/j.scienta.2019.108893
Sang Lee J, Gyu Park S, Park H et al (2002) Interaction network of human aminoacyl-tRNA synthetases and subunits of elongation factor 1 complex. Biochem Biophy Res Comm 291(1):158–164. https://doi.org/10.1006/bbrc.2002.6398
Leelatanawit R, Klanchui A, Uawisetwathana U et al (2012) Validation of reference genes for real-time PCR of reproductive system in the black tiger shrimp. PLoS ONE 7(12):1–10. https://doi.org/10.1371/journal.pone.0052677
Freitas FCP, Depintor TS, Agostini LT et al (2019) Evaluation of reference genes for gene expression analysis by real-time quantitative PCR (qPCR) in three stingless bee species (Hymenoptera: Apidae: Meliponini). Sci Rep 9(1):1–13. https://doi.org/10.1038/s41598-019-53544-0
Ridgeway JA, Timm AE (2015) Reference gene selection for quantitative real-time PCR normalization in larvae of three species of Grapholitini (Lepidoptera: Tortricidae). PLoS ONE 10(6):1–15. https://doi.org/10.1371/journal.pone.0129026
Dixit S, Jangid VK, Grover A (2019) Evaluation of suitable reference genes in Brassica juncea and its wild relative Camelina sativa for qRT-PCR analysis under various stress conditions. PLoS ONE 14(9):1–18. https://doi.org/10.1371/journal.pone.0222530
Barua B, Winkelmann DA, White HD et al (2012) Regulation of actin-myosin interaction by conserved periodic sites of tropomyosin. Pro Nat Acad Sci USA 109(45):18425–18430. https://doi.org/10.1073/pnas.1212754109
Synnergren J, Giesler T, Adak S et al (2007) Differentiating human embryonic stem cells express a unique housekeeping gene signature. Stem Cells 25:473–480. https://doi.org/10.1634/stem-cells.2006-0247
Zaffagnini M, Fermani S, Costa A et al (2013) Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci 4(450):1–18. https://doi.org/10.3389/fpls.2013.00450
de Jonge HJM, Fehrmann RSN, de Bont ESJM et al (2007) Evidence based selection of housekeeping genes. PLoS ONE 2(9):1–5. https://doi.org/10.1371/journal.pone.0000898
Niu J, Dou W, Ding T et al (2012) Evaluation of suitable reference genes for quantitative RT-PCR during development and abiotic stress in Panonychus citri (McGregor) (Acari: Tetranychidae). Mol Biol Rep 39:5841–5849. https://doi.org/10.1007/s11033-011-1394-x
Mehennaoui K, Legay S, Serchi T et al (2018) Identification of reference genes for RT-qPCR data normalization in Gammarus fossarum (Crustacea Amphipoda). Sci Rep 8(1):1–8. https://doi.org/10.1038/s41598-018-33561-1
Liu C, Xin N, Zhai Y et al (2014) Reference gene selection for quantitative real-time RT-PCR normalization in the half-smooth tongue sole (Cynoglossus semilaevis) at different developmental stages, in various tissue types and on exposure to chemicals. PLoS ONE 9(3):e91715. https://doi.org/10.1371/journal.pone.0091715
Zhou SM, Tao Z, Shen C et al (2018) β-actin gene expression is variable among individuals and not suitable for normalizing mRNA levels in Portunus trituberculatus. Fish Shellfish Immunol 81:338–342. https://doi.org/10.1016/j.fsi.2018.07.021
Kuballa A, Elizur A (2007) Novel molecular approach to study moulting in crustaceans. Bull Fish Res Agen 20:53–57
Kuballa AV, Holton TA, Paterson B et al (2011) Moult cycle specific differential gene expression profiling of the crab Portunus pelagicus. BMC Genomics 12(1):147–164. https://doi.org/10.1186/1471-2164-12-147
Gunning PW, Schevzov G, Kee AJ et al (2005) Tropomyosin isoforms: Divining rods for actin cytoskeleton function. Trends Cell Biol 15(6):333–341. https://doi.org/10.1016/j.tcb.2005.04.007
Alsailawi HA, Rosmilah M, Keong BP (2019) The effects of thermal and non-thermal treatments on protein profiles of Scylla tranquebarica (purple mud crab). Plant Arch 19:813–816
Leung NYH, Wai CYY, Shu S et al (2012) Current immunological and molecular biological perspectives on seafood allergy: A comprehensive review. Clin Rev Allergy Immunol 46(3):180–197. https://doi.org/10.1007/s12016-012-8336-9
Faber MA, Pascal M, El Kharbouchi O et al (2017) Shellfish allergens: tropomyosin and beyond. Allergy Eur J Allergy Clin Immunol 72(6):842–848. https://doi.org/10.1111/all.13115
Keenan C, Davie P, Mann D (1998b) A revision of the genus Scylla De Haan, 1833 (Crustacea: Decapoda: Brachyura: Portunidae). Raff Bull Zool 46(1):217–245
Nurul AMS, Noor ASK, Rodrigues K et al (2016) Genetic diversity of mud crabs, Scylla tranquebarica in Sabah, Malaysiabased on Cytochrome C Oxidase (COI) gene sequence. Songklanakarin J Sci Technol 38(4):365–372
Acknowledgements
The authors wish to thank the Ministry of Higher Education Malaysia (MOE) for the financial funding of this research through the Fundamental Research Grant Scheme (FRGS 2016-0085-102-02).
Author information
Authors and Affiliations
Contributions
RM and PBK conceived, designed research, analyzed data and edited manuscript. NFHA performed the experiments, analyzed data and wrote the manuscript. ZHMY edited manuscript. All authors read, commented on, and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2020_5966_MOESM1_ESM.tif
Supplementary file1 1.5% agarose gel electrophoresis showing specific qPCR products with expected size for each candidate reference genes using cDNA from S. olivacea, S. paramamosain and S. tranquebarica. Marker: GeneRuler 1 kb DNA ladder; NTC-Non-template control. (TIF 78 KB)
11033_2020_5966_MOESM2_ESM.tif
Supplementary file2 Venn diagram represented the selected most suitable reference gene for abdomen, walking leg, cheliped and across the mud crab of S. olivacea, S. paramamosain and S. tranquebarica. (TIFF 7668 KB)
Rights and permissions
About this article
Cite this article
Azemi, N.F.H., Misnan, R., Keong, P.B. et al. Reference gene and tropomyosin expression in mud crab Scylla olivacea, Scylla paramamosain and Scylla tranquebarica. Mol Biol Rep 47, 9765–9777 (2020). https://doi.org/10.1007/s11033-020-05966-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05966-7