Abstract
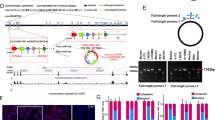
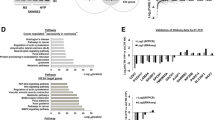
Hypoxia is associated with tumor aggressiveness and poor prognosis, including breast cancer. Low oxygen levels induces global genomic hypomethylation and hypermethylation of specific loci in tumor cells. DNA methylation is a reversible epigenetic modification, usually associated with gene silencing, contributing to carcinogenesis and tumor progression. Since the effects of DNA methyltransferase inhibitor are context-dependent and as there is little data comparing their molecular effects in normoxic and hypoxic microenvironments in breast cancer, this study aimed to understand the gene expression profiles and molecular effects in response to treatment with DNA methyltransferase inhibitor in normoxia and hypoxia, using the breast cancer model. For this, a cDNA microarray was used to analyze the changes in the transcriptome upon treatment with DNA methyltransferase inhibitor (5-Aza-2’-deoxycytidine: 5-Aza-2’-dC), in normoxia and hypoxia. Furthermore, immunocytochemistry was performed to investigate the effect of 5-Aza-2’-dC on NF-κB/p65 inflammation regulator subcellular localization and expression, in normoxia and hypoxia conditions. We observed that proinflammatory pathways were upregulated by treatment with 5-Aza-2’-dC, in both conditions. However, treatment with 5-Aza-2’-dC in normoxia showed a greater amount of overexpressed proinflammatory pathways than 5-Aza-2’-dC in hypoxia. In this sense, we observed that the NF-κB expression increased only upon 5-Aza-2’-dC in normoxia. Moreover, nuclear staining for NF-κB and NF-κB target genes upregulation, IL1A and IL1B, were also observed after 5-Aza-2’-dC in normoxia. Our results suggest that 5-Aza-2’-dC induces a greater inflammatory change, at the molecular levels, in normoxic than hypoxic tumor microenvironment. These data may support further studies and expand the understanding of the DNA methyltransferase inhibitor effects in different tumor contexts.




Similar content being viewed by others
Data availability
All data analyzed by this study are included in this article.
References
Vaupel P, Harrison L (2004) Tumor hypoxia: causative factors, compensatory mechanisms, and cellular response. Oncologist 9 Suppl 5:4–9. doi:https://doi.org/10.1634/theoncologist.9-90005-4
Bertout JA, Patel SA, Simon MC (2008) The impact of O2 availability on human cancer. Nat Rev Cancer 8(12):967–975
Höckel M, Vaupel P (2001) Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst 93(4):266–276
Fisher ER, Anderson S, Redmond C, Fisher B (1993) Pathologic findings from the national surgical adjuvant breast project protocol B-06 10-year pathologic and clinical prognostic discriminants. Cancer 71(8):2507–2514
Chi JT, Wang Z, Nuyten DSA et al (2006) Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Med. doi:https://doi.org/10.1371/journal.pmed.0030047
Keith B, Johnson RS, Simon MC (2012) HIF1 α and HIF2 α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12(1):9–22
Semenza GL (2012) Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci 33(4):207–214
Yeo CD, Kang N, Choi SY et al (2017) The role of hypoxia on the acquisition of epithelial-mesenchymal transition and cancer stemness: a possible link to epigenetic regulation. Korean J Intern Med 32:589–599
Dengler VL, Galbraith MD, Espinosa JM (2014) Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol 49(1):1–15
Bender CM, Zingg JM, Jones PA (1998) DNA methylation as a target for drug design. Pharm Res 15:175–187
Santini V, Kantarjian HM, Issa JP (2001) Changes in DNA methylation in neoplasia: Pathophysiology and therapeutic implications. Ann Intern Med 134:573–586
Thienpont B, Steinbacher J, Zhao H et al (2016) Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 537:63–68. doi:https://doi.org/10.1038/nature19081
Li H, Tong L, Tao H, Liu Z (2020) Genome-wide analysis of the hypoxia-related DNA methylation-driven genes in lung adenocarcinoma progression. Biosci Rep. doi:https://doi.org/10.1042/BSR20194200
Yoo C, Jones P (2006) Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 5:37–50. doi:https://doi.org/10.1038/nrd1930
Eden A, Gaudet F, Waghmare A, Jaenisch R (2003) Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300:2003. https://doi.org/10.1126/science.1083557
Wilson AS, Power BE, Molloy PL (2007) DNA hypomethylation and human diseases. Biochim Biophys Acta 1775:138–162. doi:https://doi.org/10.1016/j.bbcan.2006.08.007
Baylin SB (2005) DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol Oncol 2(Suppl 1):S4–S11. doi:https://doi.org/10.1038/ncponc0354
Long C, Yin B, Lu Q et al (2007) Promoter hypermethylation of the RUNX3 gene in esophageal squamous cell carcinoma. Cancer Invest 25:685–690. doi:https://doi.org/10.1080/07357900701561131
Kim JT, Li J, Song J et al (2015) Differential expression and tumorigenic function of neurotensin receptor 1 in neuroendocrine tumor cells. Oncotarget. doi:https://doi.org/10.18632/oncotarget.4745
Rawłuszko-Wieczorek AA, Horst N, Horbacka K et al (2015) Effect of DNA methylation profile on OATP3A1 and OATP4A1 transcript levels in colorectal cancer. Biomed Pharmacother. https://doi.org/10.1016/j.biopha.2015.08.026
Jones PA (1999) The DNA methylation paradox. Trends Genet 15(1):34–37
Evans IC, Barnes JL, Garner IM et al (2016) Epigenetic regulation of cyclooxygenase-2 by methylation of c8orf4 in pulmonary fibrosis. Clin Sci. doi:https://doi.org/10.1042/CS20150697
Hong X, Nelson K, Lemke N, Kalkanis SN (2012) Heparanase expression is associated with histone modifications in glioblastoma. Int J Oncol. doi:https://doi.org/10.3892/ijo.2011.1229
Chiappinelli KB, Strissel PL, Desrichard A et al (2015) Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. https://doi.org/10.1016/j.cell.2015.07.011
Roulois D, Loo Yau H, Singhania R et al (2015) DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. https://doi.org/10.1016/j.cell.2015.07.056
Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.98.1.31
Irizarry RA, Hobbs B, Collin F et al (2012) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. In: Selected Works of Terry Speed
Wang J, Vasaikar S, Shi Z et al (2017) WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 45(W1):W130–W137
Liao Y, Wang J, Jaehnig EJ et al (2019) WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. doi:https://doi.org/10.1093/nar/gkz401
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Carbon S, Ireland A, Mungall CJ et al (2009) AmiGO: online access to ontology and annotation data. Bioinformatics. https://doi.org/10.1093/bioinformatics/btn615
Carbon S, Douglass E, Dunn N et al (2019) The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. https://doi.org/10.1093/nar/gky1055
Ogata H, Goto S, Sato K et al (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27(1):29–34
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Chatterjee S, Malhotra R, Varghese F et al (2013) Quantitative immunohistochemical analysis reveals association between sodium iodide symporter and estrogen receptor expression in breast cancer. PLoS One. https://doi.org/10.1371/journal.pone.0054055
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics doi. https://doi.org/10.1093/bioinformatics/18.11.1427
Liu T, Zhang L, Joo D, Sun SC (2017) NF-κB signaling in inflammation. Signal Transduct Target Ther 2(1):1–9
Wrangle J, Wang W, Koch A et al (2013) Alterations of immune response of non-small cell lung cancer with Azacytidine. Oncotarget 4:2067–2079. doi:https://doi.org/10.18632/oncotarget.1542
Heninger E, Krueger TEG, Thiede SM et al (2016) Inducible expression of cancer-testis antigens in human prostate cancer. Oncotarget. doi:https://doi.org/10.18632/oncotarget.12711
D’anna F, Dyck L, Van Xiong J et al (2020) DNA methylation repels binding of HIF transcription factors to maintain tumour immunotolerance. bioRxiv. https://doi.org/10.1101/2020.02.07.931071
Acknowledgements
We thank to Brazilian National Cancer Institute Graduate Program in Oncology (PPGO-INCA) for the microarray hardware use, and FAPERJ, CNPq and PPGB/PROEX.
Funding
This work was funded by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (E-26/010.100944/2018), Conselho Nacional de Desenvolvimento Científico e Tecnológico (302505/2018-0) and Programa de pós-graduação em Biociências/Pró-reitoria de extensão.
Author information
Authors and Affiliations
Contributions
AI performed the experiments and wrote the manuscript draft. AI, RJ, NP, KS and PC performed the experiments and participed in analysis. FA and MA conceived the experiments and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study did not need ethical approval.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Salviano Soares de Amorim, Í., Rodrigues, J.A., Nicolau, P. et al. 5-Aza-2’-deoxycytidine induces a greater inflammatory change, at the molecular levels, in normoxic than hypoxic tumor microenvironment. Mol Biol Rep 48, 1161–1169 (2021). https://doi.org/10.1007/s11033-020-05931-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05931-4