Abstract
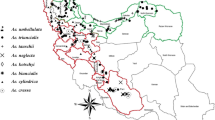
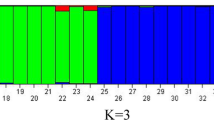
Understanding the genetic diversity and relationships between genotypes is an effective step in designing effective breeding programs. Insertional polymorphisms of retrotransposons were studied in 75 cultivated and wild grape genotypes using retrotransposon-microsatellite amplified polymorphism (REMAP) technique. In the morphological part of work, seven pomological traits with a high breeding interest were also analyzed in the cultivated genotypes. A total of 328 markers were produced by 42 primer pairs, out of which 313 markers (95.43%) were polymorphic. Number of markers ranged from 4 in loci Tvv1Fa-873, Vine1-811, Gret1Ra-855 and Tvv1Fa-890 to 12 in locus Vine1Ra-841 with an average value of 7.45. Similarity values based on Dice’s coefficient among all 75 grapevine genotypes varied from 0.41 to 0.77. Classification of genotypes using unweighted pair-group method using complete-linkage clustering led to six distinct groups. Some wild and cultivated varieties placed in the same groups. It seems there are close relationship between wild and cultivated genotypes and maybe wild genotypes are ancestor of native grapevines. Grouping of grapevine genotypes based on molecular marker data was not in agreement with clustering by agro-morphological data indicating that the most of multiplied sequences are confined to the non-coding regions of transposon elements. Results showed a substantial level of genetic diversity at molecular and pomological level and the potential of this diversity for future grape breeding programs.






Similar content being viewed by others
Data availability
Data sharing is not applicable to this article as all new created data is already contained within this article.
References
Lodhi MA, Reisch BI (1995) Nuclear DNA content of species, cultivars, and other genera of the Vitaceae. Theor Appl Genet 90:11–16
McGovern PE (2019) Ancient wine: the search for the origins of viniculture. Princeton University Press, Princeton
Amiri M (2006) The status of genetic resources of deciduous, tropical, and subtropical fruit species in Iran. In XXVII International Horticultural Congress-IHC2006: International Symposium on Asian Plants with Unique Horticultural 769, pp 159–167
Doulaty Baneh H, Grassi F, Mohammadi A, Nazemieh A, De Mattia F, Imazio S, Labra M (2007) The use of AFLP and morphological markers to study Iranian grapevine germplasm to avoid genetic erosion. J Hortic Sci Biotechnol 82:745–752
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E et al (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol 13:39
Finnegan DJ (1989) Eukaryotic transposable elements and genome evolution. Trends Genet 5:103–107
Flavell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R, Kumar A (1992) Ty1–copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res 20:3639–3644
Waugh R, McLean K, Flavell A, Pearce S, Kumar A, Thomas B, Powell W (1997) Genetic distribution of Bare–1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet 253:687–694
Flavell AJ, Knox MR, Pearce SR, Ellis TN (1998) Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J 16:643–650
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman A (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48:463–474
Wanjugi H, Coleman-Derr D, Huo N, Kianian SF, Luo M-C, Wu J, Anderson O, Gu YQ (2009) Rapid development of PCR-based genome-specific repetitive DNA junction markers in wheat. Genome 52:576–587
Mandoulakani BA, Piri Y, Darvishzadeh R, Bernoosi I, Jafari M (2012) Retroelement insertional polymorphism and genetic diversity in Medicago sativa populations revealed by IRAP and REMAP markers. Plant Mol Biol Rep 30:286–296
Kalendar R, Schulman AH (2006) IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat Protoc 1:2478
Pereira HS, Barão A, Delgado M, Morais-Cecílio L, Viegas W (2005) Genomic analysis of grapevine retrotransposon 1 (Gret1) in Vitis vinifera. Theor Appl Genet 111:871–878
Fujita K, Shimazaki M, Furiya T, Takayanagi T, Suzuki S (2009) Genetic variation among Koshu (Vitis vinifera L.) accessions generated by retrotransposon insertion into genome. Am J Enol Viticult 60:490–496
Verries C, Bes C, This P, Tesniere C (2000) Cloning and characterization of Vine-1, a LTR-retrotransposon-like element in Vitis vinifera L., and other Vitis species. Genome 43:366–376
Pelsy F, Merdinoglu D (2002) Complete sequence of Tvv1, a family of Ty1 copia-like retrotransposons of Vitis vinifera L., reconstituted by chromosome walking. Theor Appl Genet 105:614–621
Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463
Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, FitzGerald LM, Vezzulli S et al (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2:e1326
Villano C, Carputo D, Frusciante L, Santoro X, Aversano R (2014) Use of SSR and retrotransposon-based markers to interpret the population structure of native grapevines from Southern Italy. Mol Biotechnol 56(11):1011–1020
Castro I, D’Onofrio C, Martı´n JP, Marı´a Ortiz J, De Lorenzis G, Ferreira V, Pinto-Carnide O (2011) Effectiveness of AFLPs and retrotransposon-based markers for the identification of Portuguese grapevine cultivars and clones. Mol Biotechnol 52(1):26–39
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
D’Onofrio C, De Lorenzis G, Giordani T, Natali L, Cavallini A, Scalabrelli G (2010) Retrotransposon-based molecular markers for grapevine species and cultivars identification. Tree Genet Genomes 6:451–466
Rohlf J, Rohlf F, Rohlf E, Rohlf J (1993) NTSYS-pc. Numerical taxonomy and multivariate analysis system. Version 2.1. Exeter Software, Setauket, New York, NY, USA
Lanes EC, Viana JM, Paes GP, Paula MF, Maia C, Caixeta ET, Miranda GV (2014) Population structure and genetic diversity of maize inbreds derived from tropical hybrids. Genet Mol Res 13:7365–7376. https://doi.org/10.4238/2014.September.12.2
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Perrier X, Jacquemoud-Collet J (2016) DARwin software: dissimilarity analysis and representation for Windows (version 6.0. 010)
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11(1):94
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Khadivi-Khub A, Salimpour A, Rasouli M (2014) Analysis of grape germplasm from Iran based on fruit characteristics. Braz J Bot 37(2):105–113
Boz Y, Bakir M, Cerlikkol BP, Kazan K, Yilmaz F, Cakir B, Aslantas S (2011) Genetic characterization of grape (Vitis vinifera L.) germplasm from Southeast Anatolia by SSR markers. Vitis 50:99–106
Tessier C, David J, This P, Boursiquot JM, Charrier A (1999) Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor Appl Genet 98:171–177
Rold´an-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breeding 6:125–134
Fatahi R, Ebadi A, Bassil N, Mehlenbacher S, Zamani Z (2003) Characterization of Iranian grapevine cultivars using microsatellite markers. Vitis 42:185–192
Crespan M, Milani N (2001) The Muscats: a molecular analysis of synonyms, homonyms and genetic relationships within a large family of grapevine cultivars. Vitis 40:23–30
Regner F, Stadlbauer A, Eisenheld C, Kaserer H (2000) Genetic relationships among Pinots and related cultivars. Am J Enol Viticult 51:7–14
Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304:982–982
Pelsy F, Hocquigny S, Moncada X, Barbeau G, Forget D, Hinrichsen P, Merdinoglu D (2010) An extensive study of the genetic diversity within seven French wine grape variety collections. Theor Appl Genet 120:1219–1231
Forneck A (2005) Plant breeding: clonality—a concept for stability and variability during vegetative propagation. In: Luttge U et al (eds) Progress in Botany. Springer, New York, pp 164–183
Hocquigny S, Pelsy F, Dumas V, Kindt S, Heloir M, Merdinoglu D (2004) Diversification within grapevine cultivars goes through chimeric states. Genome 47:579–589
Moncada X, Pelsy F, Merdinoglu D, Hinrichsen P (2006) Genetic diversity and geographical dispersal in grapevine clones revealed by microsatellite markers. Genome 49:1459–1472
Wegscheider E, Benjak A, Forneck A (2009) Clonal variation in Pinot noir revealed by S-SAP involving universal retrotransposon-based sequences. Am J Enol Viticult 60:104–109
Benjak A, Forneck A, Casacuberta JM (2008) Genome-wide analysis of the “cut-and-paste” transposons of grapevine. PLoS ONE 3:3107
Moisy C, Garrison KE, Meredith CP, Pelsy F (2008) Characterization of ten novel Ty1/copia-like retrotransposon families of the grapevine genome. BMC Genom 9:469
Silvestroni O, Di Pietro D, Intrieri C, Vignani R, Filippetti I, Del Casino C, Scali M, Cresti M (1997) Detection of genetic diversity among clones of cv. Fortana (Vitis vinifera L.) by microsatellite DNA polymorphism analysis. Vitis 36:147–150
Frazer KA, Murray SS, Schork NJ, Topol EJ (2009) Human genetic variation and its contribution to complex traits. Nat Rev Genet 10:241–251
Tautz D, Renz M (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res 12:4127–4138
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Kidwell MG, Lisch D (1997) Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA 94:7704–7711
Laucou V, Lacombe T, Dechesne F, Siret R, Bruno J-P, Dessup M, Dessup T, Ortigosa P, Parra P et al (2011) High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor Appl Genet 122:1233–1245
Robinson J, Harding J, Vouillamoz J (2013) Wine grapes: a complete guide to 1,368 vine varieties, including their origins and flavours. Penguin Books, London
Grassi F, Labra M, Imazio S, Spada A, Sgorbati S, Scienza A, Sala F (2003) Evidence of a secondary grapevine domestication centre detected by SSR analysis. Theor Appl Genet 107:1315–1320
Snoussi H, Slimane MHB, Ruiz-García L, Martínez-Zapater JM, Arroyo-García R (2004) Genetic relationship among cultivated and wild grapevine accessions from Tunisia. Genome 47:1211–1219
Cornille A, Gladieux P, Smulders MJ, Roldan-Ruiz I, Laurens F, Le Cam B, Nersesyan A, Clavel J, Olonova M et al (2012) New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet 8:1002703
Acknowledgements
Authors thank the Support of the University of Zanjan (Iran) in the stay of Mitra Razi at CEBAS-CSIC of Murcia (Spain).
Author information
Authors and Affiliations
Contributions
MR contributed to all the experimental activities, data analysis and results explanations as well as writing the paper. MEA and RD designed and supervised the work. HDB involved in preparing the plant materials. HA contributed to analyze part of the data. PM-G and RD contributed to review and edit of manuscript. All authors contributed to the discussion. The authors accept full responsibility for the context of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
No ethical issues were promulgated.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Razi, M., Amiri, M.E., Darvishzadeh, R. et al. Assessment of genetic diversity of cultivated and wild Iranian grape germplasm using retrotransposon-microsatellite amplified polymorphism (REMAP) markers and pomological traits. Mol Biol Rep 47, 7593–7606 (2020). https://doi.org/10.1007/s11033-020-05827-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05827-3