Abstract
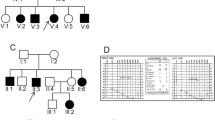
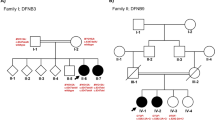
Autosomal recessive non-syndromic hearing loss (ARNSHL) is a highly heterogeneous disease, for which more than 70 genes have been identified. MYO15A mutations have been reported to cause congenital severe-to-profound HL. In this study, we applied the whole exome sequencing (WES) to find the cause of HL in an Iranian family. A proband from an Iranian non-consanguineous family with hearing impaired parents, was examined via WES, after excluding GJB2 mutations as the most common ARNSHL gene via Sanger sequencing. Co-segregation analysis of the candidate variant was done in the family members. Interpretation of variants was according to the American College of Medical Genetics and Genomics (ACMG) guidelines. WES results showed novel compound heterozygous variants (p.Arg1507Ter and p.Val2815Valfs*10) in the MYO15A gene. These two variants, residing in highly conserved regions, were found to be co-segregating in the family and fulfill the criteria of being categorized as pathogenic, according to the ACMG guidelines. Here, we report successful application of WES to identify the molecular pathogenesis of ARNSHL in a patient with ARNSHL, as an example of an extremely heterogeneous disease. In agreement with previous studies, MYO15A is regarded to be important in causing HL in Iran.




Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Mahdieh N, Rabbani B, Wiley S, Akbari MT, Zeinali S (2010) Genetic causes of nonsyndromic hearing loss in Iran in comparison with other populations. J Hum Genet 55(10):639
Babanejad M, Fattahi Z, Bazazzadegan N, Nishimura C, Meyer N, Nikzat N et al (2012) A comprehensive study to determine heterogeneity of autosomal recessive nonsyndromic hearing loss in Iran. Am J Med Genet Part A 158(10):2485–2492
Krendel M, Mooseker MS (2005) Myosins: tails (and heads) of functional diversity. Physiology. 20(4):239–251
Kalay E, Uzumcu A, Krieger E, Çaylan R, Uyguner O, Ulubil-Emiroglu M et al (2007) MYO15A (DFNB3) mutations in Turkish hearing loss families and functional modeling of a novel motor domain mutation. Am J Med Genet Part A 143(20):2382–2389
Anderson DW, Probst FJ, Belyantseva IA, Fridell RA, Beyer L, Martin DM et al (2000) The motor and tail regions of myosin XV are critical for normal structure and function of auditory and vestibular hair cells. Hum Mol Genet 9(12):1729–1738
Belyantseva IA, Boger ET, Friedman TB (2003) Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. Proc Natl Acad Sci 100(24):13958–13963
Kikkawa Y, Mburu P, Morse S, Kominami R, Townsend S, Brown SD (2004) Mutant analysis reveals whirlin as a dynamic organizer in the growing hair cell stereocilium. Hum Mol Genet 14(3):391–400
Chang MY, Lee C, Han JH, Kim MY, Park H-R, Kim N et al (2018) Expansion of phenotypic spectrum of MYO15A pathogenic variants to include postlingual onset of progressive partial deafness. BMC Med Genet 19(1):29
Sadeghian L, Tabatabaiefar MA, Fattahi N, Pourreza MR, Tahmasebi P, Alavi Z et al (2019) Next-generation sequencing reveals a novel pathological mutation in the TMC1 gene causing autosomal recessive non-syndromic hearing loss in an Iranian kindred. Int J Pediatr Otorhinolaryngol 124:99–105
Tabatabaiefar MA, Alasti F, Zohour MM, Shariati L, Farrokhi E, Farhud D et al (2011) Genetic linkage analysis of 15 DFNB loci in a group of Iranian families with autosomal recessive hearing loss. Iranian J Public Health. 40(2):34
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17(5):405
Rehman AU, Bird JE, Faridi R, Shahzad M, Shah S, Lee K et al (2016) Mutational spectrum of MYO15A and the molecular mechanisms of DFNB3 human deafness. Hum Mutat 37(10):991–1003
Hilgert N, Smith RJ, Van Camp G (2009) Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res/Rev Mutat Res. 681(2–3):189–196
Koohiyan M, Hashemzadeh-Chaleshtori M, Salehi M, Abtahi H, Reiisi S, Pourreza MR et al (2018) GJB2 mutations causing autosomal recessive non-syndromic hearing loss (ARNSHL) in two Iranian populations: report of two novel variants. Int J Pediatr Otorhinolaryngol 107:121–126
Sloan-Heggen CM, Babanejad M, Beheshtian M, Simpson AC, Booth KT, Ardalani F et al (2015) Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J Med Genet 52(12):823–829
Baux D, Vaché C, Blanchet C, Willems M, Baudoin C, Moclyn M et al (2017) Combined genetic approaches yield a 48% diagnostic rate in a large cohort of French hearing-impaired patients. Sci Rep. 7(1):1–10
Bashir R, Fatima A, Naz S (2012) Prioritized sequencing of the second exon of MYO15A reveals a new mutation segregating in a Pakistani family with moderate to severe hearing loss. Eur J Med Genet. 55(2):99–102
Shearer AE, Hildebrand MS, Webster JA, Kahrizi K, Meyer NC, Jalalvand K et al (2009) Mutations in the first MyTH4 domain of MYO15A are a common cause of DFNB3 hearing loss. Laryngoscope. 119(4):727–733
Fattahi Z, Shearer AE, Babanejad M, Bazazzadegan N, Almadani SN, Nikzat N et al (2012) Screening for MYO15A gene mutations in autosomal recessive nonsyndromic, GJB2 negative Iranian deaf population. Am J Med Genet Part A 158(8):1857–1864
Lane H (2005) Ethnicity, ethics, and the deaf-world. J Deaf Stud Deaf Educ. 10(3):291–310
Zhang J, Guan J, Wang H, Yin L, Wang D, Zhao L et al (2019) Genotype-phenotype correlation analysis of MYO15A variants in autosomal recessive non-syndromic hearing loss. BMC Med Genet 20(1):60
Cengiz FB, Duman D, Sırmacı A, Tokgöz-Yilmaz S, Erbek S, Öztürkmen-Akay H et al (2010) Recurrent and private MYO15A mutations are associated with deafness in the Turkish population. Genet Test Mol Biomark. 14(4):543–550
Fang Q, Indzhykulian AA, Mustapha M, Riordan GP, Dolan DF, Friedman TB et al (2015) The 133-kDa N-terminal domain enables myosin 15 to maintain mechanotransducing stereocilia and is essential for hearing. Elife. 4:e08627
Nal N, Ahmed ZM, Erkal E, Alper ÖM, Lüleci G, Dinç O et al (2007) Mutational spectrum of MYO15A: the large N-terminal extension of myosin XVA is required for hearing. Hum Mutat 28(10):1014–1019
Abedi A, Rostami M, Abedi S, Sudmand N, Movallali G (2018) Marital satisfaction in deaf couples: a review study. Audit Vestib Res
Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Rayyan AA et al (2011) Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol 12(9):R89
Vozzi D, Morgan A, Vuckovic D, D’Eustacchio A, Abdulhadi K, Rubinato E et al (2014) Hereditary hearing loss: a 96 gene targeted sequencing protocol reveals novel alleles in a series of Italian and Qatari patients. Gene 542(2):209–216
Park JH, Kim NK, Kim AR, Rhee J, Oh SH, Koo J-W et al (2014) Exploration of molecular genetic etiology for Korean cochlear implantees with severe to profound hearing loss and its implication. Orphanet J Rare Dis. 9(1):167
Neveling K, Feenstra I, Gilissen C, Hoefsloot LH, Kamsteeg EJ, Mensenkamp AR et al (2013) A post hoc comparison of the utility of S anger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat 34(12):1721–1726
Miyagawa M, Nishio S-Y, Hattori M, Moteki H, Kobayashi Y, Sato H et al (2015) Mutations in the MYO15A Gene are a significant cause of nonsyndromic hearing loss: massively parallel DNA sequencing–based analysis. Ann Otol Rhinol Laryngol. 124(1_suppl):158S–168S
Sommen M, Schrauwen I, Vandeweyer G, Boeckx N, Corneveaux JJ, van den Ende J et al (2016) DNA diagnostics of hereditary hearing loss: a targeted resequencing approach combined with a mutation classification system. Hum Mutat 37(8):812–819
Vona B, Müller T, Nanda I, Neuner C, Hofrichter MA, Schröder J et al (2014) Targeted next-generation sequencing of deafness genes in hearing-impaired individuals uncovers informative mutations. Genet Med. 16(12):945–953
Moteki H, Azaiez H, Booth KT, Shearer AE, Sloan CM, Kolbe DL et al (2016) Comprehensive genetic testing with ethnic-specific filtering by allele frequency in a Japanese hearing-loss population. Clin Genet 89(4):466–472
Liburd N, Ghosh M, Riazuddin S, Naz S, Khan S, Ahmed Z et al (2001) Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum Genet 109(5):535–541
Li W, Guo L, Li Y, Wu Q, Li Q, Li H et al (2016) A novel recessive truncating mutation in MYO15A causing prelingual sensorineural hearing loss. Int J Pediatr Otorhinolaryngol 81:92–95
Bai X, Nian S, Feng L, Ruan Q, Luo X, Wu M et al (2019) Identification of novel variants in MYO15A, OTOF, and RDX with hearing loss by next-generation sequencing. Mol Genet Genomic Med. 7(8):e808
Belguith H, Aifa-Hmani M, Dhouib H, Said MB, Mosrati MA, Lahmar I et al (2009) Screening of the DFNB3 locus: identification of three novel mutations of MYO15A associated with hearing loss and further suggestion for two distinctive genes on this locus. Genet Test Mol Biomark. 13(1):147–151
Ammar-Khodja F, Bonnet C, Dahmani M, Ouhab S, Lefèvre GM, Ibrahim H et al (2015) Diversity of the causal genes in hearing impaired Algerian individuals identified by whole exome sequencing. Mol Genet Genomic Med. 3(3):189–196
Duman D, Sirmaci A, Cengiz FB, Ozdag H, Tekin M (2011) Screening of 38 genes identifies mutations in 62% of families with nonsyndromic deafness in Turkey. Genet Test Mol Biomark. 15(1–2):29–33
Chang MY, Kim AR, Kim NK, Lee C, Lee KY, Jeon W-S et al (2015) Identification and clinical implications of novel MYO15A mutations in a non-consanguineous Korean family by targeted exome sequencing. Mol Cells 38(9):781
Yang T, Wei X, Chai Y, Li L, Wu H (2013) Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis. 8(1):85
Shahin H, Rahil M, Rayan AA, Avraham KB, King M-C, Kanaan M et al (2010) Nonsense mutation of the stereociliar membrane protein gene PTPRQ in human hearing loss DFNB84. J Med Genet 47(9):643–645
Zarepour N, Koohiyan M, Taghipour-Sheshdeh A, Nemati-Zargaran F, Saki N, Mohammadi-Asl J et al (2019) Identification and clinical implications of a novel MYO15A variant in a consanguineous Iranian family by targeted exome sequencing. Audiol Neurotol. 24(1):25–31
Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW et al (1998) Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 280(5368):1447–1451
Woo H-M, Park H-J, Baek J-I, Park M-H, Kim U-K, Sagong B et al (2013) Whole-exome sequencing identifies MYO15A mutations as a cause of autosomal recessive nonsyndromic hearing loss in Korean families. BMC Med Genet 14(1):72
Budde BS, Aly MA, Mohamed MR, Breß A, Altmüller J, Motameny S et al (2020) Comprehensive molecular analysis of 61 Egyptian families with hereditary nonsyndromic hearing loss. Clin Genet
Xia H, Huang X, Guo Y, Hu P, He G, Deng X et al (2015) Identification of a novel MYO15A mutation in a Chinese family with autosomal recessive nonsyndromic hearing loss. PLoS ONE 10(8):e0136306
Schrauwen I, Sommen M, Corneveaux JJ, Reiman RA, Hackett NJ, Claes C et al (2013) A sensitive and specific diagnostic test for hearing loss using a microdroplet PCR-based approach and next generation sequencing. Am J Med Genet Part A 161(1):145–152
Akbariazar E, Vahabi A, Abdi Rad I (2019) Report of a novel splicing mutation in the MYO15A gene in a patient with sensorineural hearing loss and spectrum of the MYO15A mutations. Clin Med Insights 12:1179547619871907
Lezirovitz K, Pardono E, de MelloAuricchio MT, e Silva FLDC, Lopes JJ, Abreu-Silva RS et al (2008) Unexpected genetic heterogeneity in a large consanguineous Brazilian pedigree presenting deafness. Eur J Hum Genet 16(1):89–96
Acknowledgements
We take this opportunity to express our special thanks to the staff at the Isfahan Cochlear Implantation Center and also to the patient and family.
Funding
This work was financially supported by Isfahan University of Medical Sciences Grant Nos. 295176, 396133 and 394805.
Author information
Authors and Affiliations
Contributions
Study design: MAT; Enrolling patient and clinical data collection: HA; Data collection, analysis, and interpretation: MAT, AS, SN1, SN2 and ZN; manuscript preparation: AS, SN1, ZN; critically reviewed by MAT. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study was approved by the Review Board of Isfahan University of Medical Sciences (Grant nos. 295176, 396133 and 394805).
Informed consent
Written informed consent was obtained from all of the participants in the study and a written consent to participate was obtained from the parents of the patient (younger than the age of 16). Written informed consent for publication of clinical details and clinical images was obtained from the all of the participants and from the parents the participant under the age of 18.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sarmadi, A., Nasrniya, S., Narrei, S. et al. Whole exome sequencing identifies novel compound heterozygous pathogenic variants in the MYO15A gene leading to autosomal recessive non-syndromic hearing loss. Mol Biol Rep 47, 5355–5364 (2020). https://doi.org/10.1007/s11033-020-05618-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05618-w