Abstract
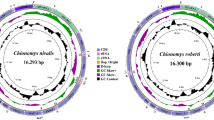
The speckled wood-pigeon, Columba hodgsonii, is mainly distributed in Bhutan, China, India, Laos, Myanmar, Nepal, Pakistan, and Thailand. Although there are several studies on birds in the family Columbidae, no study has focused on C. hodgsonii, a member of this family. Therefore, this study aimed to clarify the phylogenetic status of C. hodgsonii. The complete mitochondrial genome (mitogenome) of C. hodgsonii was sequenced and characterized and compared with those of other Columba species. The C. hodgsonii mitogenome was found to be 17,477 bp in size and contained 13 PCGs, two rRNAs, 22 tRNAs, and one CR. Of the 37 genes encoded by the C. hodgsonii mitogenome, 28 were on the heavy strand and nine were on the light strand. Twelve PCGs were initiated by ATN codons and one PCG harbored an incomplete termination codon (T-). The base composition of C. hodgsonii PCGs was A = 29.44%, T = 24.37%, G = 12.43%, and C = 33.76%. For the whole mitogenome, including PCGs, rRNAs, tRNAs, and the control region, the AT-skew was positive, and the GC-skew was negative. Phylogenetic analysis based on the base sequences of 13 PCGs from 28 Columbidae species and one outgroup using maximum likelihood and Bayesian inference indicated that C. hodgsonii belongs to the genus Columba and that the family Columbidae is monophyletic.





Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21(9):2104–2105. https://doi.org/10.1093/bioinformatics/bti263
Batalha-Filho H, Irestedt M, Fjeldså J, Ericson PG, Silveira LF, Miyaki CY (2013) Molecular systematics and evolution of the Synallaxis ruficapilla complex (Aves: Furnariidae) in the Atlantic Forest. Mol Phylogenet Evol 67(1):86–94. https://doi.org/10.1016/j.ympev.2013.01.007
Bensch S (2000) Mitochondrial genomic rearrangements in songbirds. Mol Biol Evol 17(1):107–113. https://doi.org/10.1093/oxfordjournals.molbev.a026223
Biewener AA, Dial KP (2010) In vivo strain in the humerus of pigeons (columba livia) during flight. J Morphol 225(1):61–75. https://doi.org/10.1002/jmor.1052250106
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27(8):1767–1780. https://doi.org/10.1093/nar/27.8.1767
Bruxaux J, Gabrielli M, Ashari H, Prŷs-Jones R, Joseph L, MilÁ B (2018) Recovering the evolutionary history of crowned pigeons (columbidae: goura ): implications for the biogeography and conservation of new guinean lowland birds. Mol Phylogenet Evol 120(5):248. https://doi.org/10.1016/j.ympev.2017.11.022
Davies NB (1978) Territorial defence in the speckled wood butterfly (pararge aegeria): the resident always wins. Anim Behav 26:138–147. https://doi.org/10.1016/0003-3472(78)90013-1
DuBay SG, Witt CC (2012) An improved phylogeny of the Andean tit-tyrants (Aves, Tyrannidae): more characters trump sophisticated analyses. Mol Phylogenet Evol 64(2):285–296. https://doi.org/10.1016/j.ympev.2012.04.002
Escalante AA (2011) Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Mol Biol Evol 28(6):1927–1942. https://doi.org/10.1093/molbev/msr014
Haring E, Kruckenhauser L, Gamauf A, Riesing MJ, Pinsker W (2001) The complete sequence of the mitochondrial genome of Buteo buteo (Aves, Accipitridae) indicates an early split in the phylogeny of raptors. Mol Biol Evol 18(10):1892–1904. https://doi.org/10.1093/oxfordjournals.molbev.a003730
Heth CD, Kelly DM, Spetch ML (1998) Pigeons’ (columba livia) encoding of geometric and featural properties of a spatial environment. J Comp Psychol 112(3):259–269. https://doi.org/10.1037/0735-7036.112.3.259
Hua R, Cui D, Liu J, You Y, Jia T (2018) Next-generation sequencing yields the complete mitogenome of red-crowned crane (G. japonensis). Mitochondrial DNA Part B 3(2):556–557. https://doi.org/10.1080/23802359.2018.1467220
Huang Z, Tu F, Liu X (2016) Determination of the complete mitogenome of Spotted Dove, Spilopelia chinensis (Columbiformes: Columbidae). Mitochondrial DNA Part A 27(6):4224–4225. https://doi.org/10.3109/19401736.2015.1022750
Huang Z, Tu F, Murphy RW (2016) Analysis of the complete mitogenome of Oriental turtle dove (Streptopelia orientalis) and implications for species divergence. Biochem Syst Ecol 65:209–213. https://doi.org/10.1016/j.bse.2016.02.022
Jang KH, Ryu SH, Kang SG, Hwang UW (2016) Complete mitochondrial genome of the Japanese wood pigeon, Columba janthina janthina (Columbiformes, Columbidae). Mitochondrial DNA A 27(3):2165–2166. https://doi.org/10.3109/19401736.2014.982608
Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14(6):587–589. https://doi.org/10.1038/nmeth.4285
Kan X, Yuan J, Zhang L, Li X, Yu L, Chen L, Yang J (2013) Complete mitochondrial genome of the Tristram's Bunting, Emberiza tristrami (Aves: Passeriformes): the first representative of the family Emberizidae with six boxes in the central conserved domain II of control region. Mitochondrial DNA 24(6):648–650. https://doi.org/10.3109/19401736.2013.772165
Kan XZ, Li XF, Lei ZP, Wang M, Chen L, Gao H, Yang ZY (2010) Complete mitochondrial genome of Cabot’s tragopan, Tragopan caboti (Galliformes: Phasianidae). Genet Mol Res 9(2):1204–1216. https://doi.org/10.4238/vol9-2gmr820
Kan XZ, Li XF, Zhang LQ, Chen L, Qian CJ, Zhang XW, Wang L (2010) Characterization of the complete mitochondrial genome of the Rock pigeon, Columba livia (Columbiformes: Columbidae). Genet Mol Res 9(2):1234–1249. https://doi.org/10.4238/vol9-2gmr853
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054+
Lim HC, Zou F, Taylor SS, Marks BD, Moyle RG, Voelker G, Sheldon FH (2010) Phylogeny of magpie-robins and shamas (Aves: Turdidae: Copsychus and Trichixos): implications for island biogeography in Southeast Asia. J Biogeogr 37(10):1894–1906. https://doi.org/10.1111/j.1365-2699.2010.02343.x
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44(W1):W54–W57. https://doi.org/10.1093/nar/gkw413
Mindell DP, Sorenson MD, Dimcheff DE (1998) Multiple independent origins of mitochondrial gene order in birds. Proc Natl Acad Sci USA 95(18):10693–10697. https://doi.org/10.1073/pnas.95.18.10693
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274. https://doi.org/10.1093/molbev/msu300
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41(3):353–358. https://doi.org/10.1007/bf00186547
Pombert JF, Otis C, Lemieux C, Turmel M (2004) The complete mitochondrial DNA sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) highlights distinctive evolutionary trends in the Chlorophyta and suggests a sister-group relationship between the Ulvophyceae and Chlorophyceae. Mol Biol Evol 21(5):922–935. https://doi.org/10.1093/molbev/msh099
Rambaut A (2007). FigTree, a graphical viewer of phylogenetic trees. https://tree.bio.ed.ac.uk/software/figtree. Accessed 24 Jul 2015
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542. https://doi.org/10.1093/sysbio/sys029
Slack KE, Janke A, Penny D, Arnason U (2003) Two new avian mitochondrial genomes (penguin and goose) and a summary of bird and reptile mitogenomic features. Gene 302(1–2):43–52. https://doi.org/10.1016/s0378111902010533
Soares AER, Novak BJ, Haile J, Heupink TH (2016) Complete mitochondrial genomes of living and extinct pigeons revise the timing of the columbiform radiation. BMC Evol Biol 16(1):230. https://doi.org/10.1186/s12862-016-0800-3
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinformatics 21(4):537–539. https://doi.org/10.1093/bioinformatics/bti054
Sun CH, Liu B, Lu CH (2019) Complete mitochondrial genome of the Siberian thrush, Geokichla sibirica sibirica (Aves, Turdidae). Mitochondrial DNA Part B 4(1):1150–1151. https://doi.org/10.1080/23802359.2019.1591195
Sun CH, Liu HY, Lu CH (2020) Five new mitogenomes of Phylloscopus (Passeriformes, Phylloscopidae): Sequence, structure, and phylogenetic analyses. Int J Biol Macromol 146:638–647. https://doi.org/10.1016/j.ijbiomac.2019.12.253
Wilson RT, Lewis JG (2008) Observations on the speckled pigeon columba guinea in tigrai, ethiopia. Ibis 119(2):195–198. https://doi.org/10.1111/j.1474-919X.1977.tb03537.x
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141:173–216. https://doi.org/10.1016/S0074-7696(08)62066-5
Yamaoka K, Nakagawa T, Uno T (1978) Application of Akaike's information criterion (AIC) in the evaluation of linear pharmacokinetic equations. J Pharmacokinet Biopharm 6(2):165–175. https://doi.org/10.1007/BF01117450
Yang QQ, Liu SW, Song F, Liu GF, Yu XP (2018) Comparative mitogenome analysis on species of four apple snails (Ampullariidae: Pomacea). Int J Biol Macromol 118:525–533. https://doi.org/10.1016/j.ijbiomac.2018.06.092
Zhang T, Zhang GX, Han KP, Tang Y, Wang JY, Fan QC, Wang YJ (2015) Molecular cloning and characterization, and prokaryotic expression of the GnRH1 gene obtained from Jinghai yellow chicken. Genet Mol Res 14(1):2831–2849. https://doi.org/10.4238/2015.March.31.14
Acknowledgements
This study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This study was approved by the ethics committee of Nanjing Forestry University. Muscle tissue used here was collected from a dead bird. No live animals were involved in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, HY., Sun, CH., Zhu, Y. et al. Complete mitogenomic and phylogenetic characteristics of the speckled wood-pigeon (Columba hodgsonii). Mol Biol Rep 47, 3567–3576 (2020). https://doi.org/10.1007/s11033-020-05448-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05448-w