Abstract
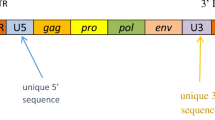
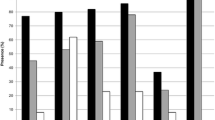
Although multiple sclerosis (MS) is one of the most common central nervous system diseases in young adults, little is known about its etiology. Several human endogenous retroviruses (ERVs) are considered to play a role in MS. We are interested in which ERVs can be identified in the vicinity of MS associated genetic marker to find potential initiators of MS. We analysed the chromosomal regions surrounding 58 single nucleotide polymorphisms (SNPs) that are associated with MS identified in one of the last major genome wide association studies. We scanned these regions for putative endogenous retrovirus sequences with large open reading frames (ORFs). We observed that more retrovirus-related putative ORFs exist in the relatively close vicinity of SNP marker indices in multiple sclerosis compared to control SNPs. We found very high homologies to HERV-K, HCML-ARV, XMRV, Galidia ERV, HERV-H/env62 and XMRV-like mouse endogenous retrovirus mERV-XL. The associated genes (CYP27B1, CD6, CD58, MPV17L2, IL12RB1, CXCR5, PTGER4, TAGAP, TYK2, ICAM3, CD86, GALC, GPR65 as well as the HLA DRB1*1501) are mainly involved in the immune system, but also in vitamin D regulation. The most frequently detected ERV sequences are related to the multiple sclerosis-associated retrovirus, the human immunodeficiency virus 1, HERV-K, and the Simian foamy virus. Our data shows that there is a relation between MS associated SNPs and the number of retroviral elements compared to control. Our data identifies new ERV sequences that have not been associated with MS, so far.



Similar content being viewed by others
References
Sadovnick AD, Ebers GC (1993) Epidemiology of multiple sclerosis: a critical overview. Can J Neurol Sci 20:17–29
Islam T, Gauderman WJ, Cozen W, Mack TM (2007) Childhood sun exposure influences risk of multiple sclerosis in monozygotic twins. Neurology 69:381–388
Ebers GC, Sadovnick AD, Risch NJ (1995) A genetic basis for familial aggregation in multiple sclerosis. Nature 377:150–151
Pender MP, Greer JM (2007) Immunology of multiple sclerosis. Curr Allergy Asthma Rep 7:285–292
Emmer A, Staege MS, Kornhuber ME (2014) The retrovirus/superantigen hypothesis of multiple sclerosis. Cell Mol Neurobiol 34:1087–1096
Tselis A (2011) Evidence for viral etiology of multiple sclerosis. Semin Neurol 31:307–316
Jern P, Coffin JM (2008) Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732
Dupressoir A, Lavialle C, Heidmann T (2012) From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33:663–671
Oja M, Peltonen J, Blomberg J, Kaski S (2007) Methods for estimating human endogenous retrovirus activities from EST databases. BMC Bioinform 8(Suppl 2):11
Dolei A (2006) Endogenous retroviruses and human disease. Expert Rev Clin Immunol 2:149–167
Perron H, Hamdani N, Faucard R, Lajnef M, Jamain S, Daban-Huard C et al (2012) Molecular characteristics of human endogenous retrovirus type-W in schizophrenia and bipolar disorder. Transl psychiatry 2:e201
Mason MJ, Speake C, Gersuk VH, Nguyen QA, O’Brien KK, Odegard JM et al (2014) Low HERV-K (C4) copy number is associated with type 1 diabetes. Diabetes 63:1789–1795
Goering W, Schmitt K, Dostert M, Schaal H, Deenen R, Mayer J et al (2015) Human endogenous retrovirus HERV-K (HML-2) activity in prostate cancer is dominated by a few loci. Prostate 75:1958–1971
De la Hera B, Varadé J, García-Montojo M, Alcina A, Fedetz M, Alloza I et al (2014) Human endogenous retrovirus HERV-Fc1 association with multiple sclerosis susceptibility: a meta-analysis. PLoS One 9:e90182
Christensen T (2010) HERVs in neuropathogenesis. J Neuroimmune Pharmacol 5:326–335
Nexø BA, Christensen T, Frederiksen J, Møller-Larsen A, Oturai AB, Villesen P et al (2011) The etiology of multiple sclerosis: genetic evidence for the involvement of the human endogenous retrovirus HERV-Fc1. PLoS One 6:e16652
Nellåker C, Yao Y, Jones-Brando L, Mallet F, Yolken RH, Karlsson H (2006) Transactivation of elements in the human endogenous retrovirus W family by viral infection. Retrovirology 3:44
Sutkowski N, Conrad B, Thorley-Lawson DA, Huber BT (2001) Epstein–Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 15:579–589
Levin LI, Munger KL, O’Reilly EJ, Falk KI, Ascherio A (2010) Primary infection with the Epstein–Barr virus and risk of multiple sclerosis. Ann Neurol 67:824–830
Kurth R, Bannert N (2010) Beneficial and detrimental effects of human endogenous retroviruses. Int J Cancer 126:306–314
Landry JR, Mager DL (2003) Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J Virol 77:7459–7466
Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E (2006) At least 50% of human-specific HERV-K(HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol 80:10752–10762
Clausen J (2003) Endogenous retroviruses and MS: using ERVs as disease markers. Int MS J MS Forum 10:22–28
Matthews AG, Finkelstein DM, Betensky RA (2008) Analysis of familial aggregation studies with complex ascertainment schemes. Stat Med 27:5076–5092
Nielsen NM, Westergaard T, Rostgaard K, Frisch M, Hjalgrim H, Wohlfahrt J et al (2005) Familial risk of multiple sclerosis: a nationwide cohort study. Am J Epidemiol 162:774–778
Sadovnick AD, Armstrong H, Rice GPA, Bulman D, Hashimoto L, Party DW et al (1993) A population-based study of multiple sclerosis in twins: update. Ann Neurol 33:281–285
Dankowski T, Buck D, Andlauer TF, Antony G, Bayas A, Bechmann L et al (2015) Successful replication of GWAS hits for multiple sclerosis in 10,000 Germans using the exome array. Genet Epidemiol 39:601–608
International Multiple Sclerosis Genetics Consortium, and Wellcome Trust Case Control Consortium 2 (2011) Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476:214–219
Chao MJ, Barnardo MC, Lincoln MR, Ramagopalan SV, Herrera BM, Dyment DA et al (2008) HLA class I alleles tag HLA-DRB1* 1501 haplotypes for differential risk in multiple sclerosis susceptibility. Proc Natl Acad Sci 105:13069–13074
Greshake B, Bayer PE, Rausch H, Reda J (2014) OpenSNP—a crowdsourced web resource for personal genomics. PLoS One 9:e89204
Rice P, Longden I, Bleasby A (2000) EMBOSS: The European molecular biology open software suite. Trends Genet 16:276–277
Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA et al (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109
Smit AFA, Hubley R, Green P (1996) RepeatMasker Open-3.0
Tönjes RR, Löwer R, Boller K, Denner J, Hasenmaier B, Kirsch H et al (1996) HERV-K: the biologically most active human endogenous retrovirus family. JAIDS 13:261–267
Bhardwaj N, Maldarelli F, Mellors J, Coffin JM (2014) HIV-1 infection leads to increased transcription of human endogenous retrovirus HERV-K (HML-2) proviruses in vivo but not to increased virion production. J Virol 88:11108–11120
Krishnamurthy J, Rabinovich BA, Mi T, Switzer KC, Olivares S, Maiti SN et al (2015) Genetic engineering of T cells to target HERV-K, an ancient retrovirus on melanoma. Clin Cancer Res 21:3241–3251
Sicat J, Sutkowski N, Huber BT (2005) Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J Rheumatol 32:1821–1831
Tai AK, O’Reilly EJ, Alroy KA, Simon KC, Munger KL, Huber BT et al (2008) Human endogenous retrovirus-K18 Env as a risk factor in multiple sclerosis. Mult Scler 14:1175–1180
Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J (2001) Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol 11:1531–1535
Krzysztalowska-Wawrzyniak M, Ostanek M, Clark J, Binczak-Kuleta A, Ostanek L, Kaczmarczyk M et al (2011) The distribution of human endogenous retrovirus K-113 in health and autoimmune diseases in Poland. Rheumatology 50:1310–1314
Moyes DL, Goris A, Ban M, Compston A, Griffiths DJ, Sawcer S et al (2008) HERV-K113 is not associated with multiple sclerosis in a large family-based study. AIDS Res Hum Retrovir 24:363–365
Naveira H, Bello X, Abal-Fabeiro JL, Maside X (2014) Evidence for the persistence of an active endogenous retrovirus (ERVE) in humans. Genetica 142:451–460
Prusty BK, zur Hausen H, Schmidt R, Kimmel R, de Villiers EM (2008) Transcription of HERV-E and HERV-E-related sequences in malignant and non-malignant human haematopoietic cells. Virology 382:37–45
Schlaberg R, Choe DJ, Brown KR, Thaker HM, Singh IR (2009) XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci 106:16351–16356
Maric R, Pedersen FS, Kjeldbjerg A, Moeller-Larsen A, Bahrami S, Brudek T et al (2011) Absence of xenotropic murine leukaemia virus-related virus in Danish patients with multiple sclerosis. Retrovirology 8:1
Mendoza R, Vaughan AE, Miller AD (2011) The left half of the XMRV retrovirus is present in an endogenous retrovirus of NIH/3T3 Swiss mouse cells. J Virol 85:9247–9248
Sato E, Furuta RA, Miyazawa T (2010) An endogenous murine leukemia viral genome contaminant in a commercial RT-PCR kit is amplified using standard primers for XMRV. Retrovirology 7:110
Niewiadomska AM, Gifford RJ (2013) The extraordinary evolutionary history of the reticuloendotheliosis viruses. PLoS Biol 11(8):e1001642
Wilkinson DA, Mager DL, Leong J-AC (1994) Endogenous human retroviruses. In: Levy AJ (ed) The retroviridae. Springer, Boston, pp 465–535
de Parseval N, Casella JF, Gressin L, Heidmann T (2001) Characterization of the three HERV-H proviruses with an open envelope reading frame encompassing the immunosuppressive domain and evolutionary history in primates. Virology 279:558–569
Brudek T, Christensen T, Aagaard L, Petersen T, Hansen HJ, Møller-Larsen A (2009) B cells and monocytes from patients with active multiple sclerosis exhibit increased surface expression of both HERV-H Env and HERV-W Env, accompanied by increased seroreactivity. Retrovirology 6:104
Jovičić S, Ignjatović S, Majkić-Singh N (2012) Biochemistry and metabolism of vitamin D/Biohemija i metabolizam vitamina D. J Med Biochem 31:309–315
Dalla Rosa I, Durigon R, Pearce SF, Rorbach J, Hirst EM, Vidoni S et al (2014) MPV17L2 is required for ribosome assembly in mitochondria. Nucleic Acids Res 42:8500–8515
Presky DH, Yang H, Minetti LJ, Chua AO, Nabavi N, Wu CY et al (1996) A functional interleukin 12 receptor complex is composed of two β-type cytokine receptor subunits. Proc Natl Acad Sci 93:14002–14007
Takahashi N, Akahoshi M, Matsuda A, Ebe K, Inomata N, Obara K et al (2005) Association of the IL12RB1 promoter polymorphisms with increased risk of atopic dermatitis and other allergic phenotypes. Hum Mol Genet 14:3149–3159
Dobner T, Wolf I, Emrich T, Lipp M (1992) Differentiation-specific expression of a novel G protein-coupled receptor from Burkitt’s lymphoma. Eur J Immunol 22:2795–2799
Müller G, Lipp M (2001) Signal transduction by the chemokine receptor CXCR5: structural requirements for G protein activation analyzed by chimeric CXCR1/CXCR5 molecules. Biol Chem 382:1387–1397
Bürkle A, Niedermeier M, Schmitt-Gräff A, Wierda WG, Keating MJ, Burger JA (2007) Overexpression of the CXCR5 chemokine receptor, and its ligand, CXCL13 in B-cell chronic lymphocytic leukemia. Blood 110:3316–3325
Mao M, Biery MC, Kobayashi SV, Ward T, Schimmack G, Burchard J et al (2004) T lymphocyte activation gene identification by coregulated expression on DNA microarrays. Genomics 83:989–999
Connelly TM, Sehgal R, Berg AS, Hegarty JP, Deiling S, Stewart DB et al (2012) Mutation in TAGAP is protective of anal sepsis in ileocolic Crohn’s disease. Dis Colon Rectum 55:1145–1152
Graham DC, Morris DL, Bhangale TR, Criswell LA, Syvanen AC, Ronnblom L et al (2011) Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet 7:e1002341
Fawcett J, Holness CL, Needham LA, Turley H, Gattert KC, Mason DY et al (1992) Molecular cloning of ICAM-3, a third ligand for LFA-1, constitutively expressed on resting leukocytes. Nature 360:481–484
Del Pozo MA, Cabañas C, Montoya MC, Ager A, Sánchez-Mateos P, Sánchez-Madrid F (1997) ICAMs redistributed by chemokines to cellular uropods as a mechanism for recruitment of T lymphocytes. J Cell Biol 137:493–508
Gregory CD, Devitt A, Moffatt O (1998) Roles of ICAM-3 and CD14 in the recognition and phagocytosis of apoptotic cells by macrophages. Biochem Soc Trans 26:644–649
Vilella R, Mila J, Lozano F, Alberola-Ila J, Places L, Vives J (1990) Involvement of the CDw50 molecule in allorecognition. Tissue Antigens 36:203–210
Ramagopalan SV, Maugeri NJ, Handunnetthi L, Lincoln MR, Orton SM, Dyment DA et al (2009) Expression of the multiple sclerosis-associated MHC class II Allele HLA-DRB1* 1501 is regulated by vitamin D. PLoS Genet 5:e1000369
Laufer G, Mayer J, Mueller BF, Mueller-Lantzsch N, Ruprecht K (2009) Analysis of transcribed human endogenous retrovirus W env loci clarifies the origin of multiple sclerosis-associated retrovirus env sequences. Retrovirology 6:4690–4696
Durán E, Gálvez J, Patrignani G, Izquierdo G (2004) Multiple sclerosis-like illness in a HIV-1 patient. J Neurol 251:1142–1144
Chin JH (2015) Multiple sclerosis and HIV-1 infection: case report of a HIV controller. J Neurovirol 21:1–4
Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, Cookinham S, King SR, Markovitz DM (2012) Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J Virol 86:7790–7805
Mouinga-Ondémé A, Caron M, Nkoghé D, Telfer P, Marx P, Kazanji M (2012) Cross-species transmission of simian foamy virus to humans in rural Gabon, Central Africa. J Virol 86:1255–1260
Mouinga-Ondémé A, Kazanji M (2013) Simian foamy virus in non-human primates and cross-species transmission to humans in Gabon: an emerging zoonotic disease in central Africa? Viruses 5:1536–1552
Luzi P, Rafi MA, Wenger DA (1995) Structure and organization of the human galactocerebrosidase (GALC) gene. Genomics 26:407–409
Alexander SP, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M et al (2013) The concise guide to pharmacology 2013/14: G protein-coupled receptors. Br J Pharmacol 170:1459–1581
Ihara Y, Kihara Y, Hamano F, Yanagida K, Morishita Y, Kunita A et al (2010) The G protein-coupl ed receptor T-cell death-associated gene 8 (TDAG8) facilitates tumor development by serving as an extracellular pH sensor. Proc Natl Acad Sci 107:17309–17314
Griffiths DJ (2001) Endogenous retroviruses in the human genome sequence. Genome Biol 2:10171–10175
McCLINTOCK B (1956) Controlling elements and the gene. In: Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor Laboratory Press, pp 197–216
Ohno S (1972) So much “junk” DNA in our genome. In: Brookhaven Symposium in Biology, pp 366–370
Warren IA, Naville M, Chalopin D, Levin P, Berger CS, Galiana D et al (2015) Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res 23:1–27
Volkman HE, Stetson DB (2014) The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol 15:415–422
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Brütting, C., Emmer, A., Kornhuber, M. et al. A survey of endogenous retrovirus (ERV) sequences in the vicinity of multiple sclerosis (MS)-associated single nucleotide polymorphisms (SNPs). Mol Biol Rep 43, 827–836 (2016). https://doi.org/10.1007/s11033-016-4004-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-4004-0