Abstract
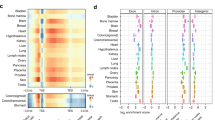
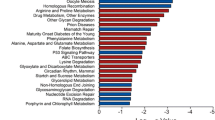
Mastermind-like 1 (MAML1) is a transcriptional coregulator that has been associated with early development of many systems such as neuronal, muscular and urogenital. The present study aimed to explore the genome wide effects of MAML1 on DNA methylation and RNA expression in human embryonic kidney cells. Infinium HumanMethylation450 BeadChip Illumina array, methylation-sensitive high-resolution melt technique, Chip Analysis Methylation Pipeline and RNA profiling approaches were used to study MAML1 effects on the epigenome. We found that 11802 CpG sites were differentially methylated in MAML1-expressing cells while only 225 genes were differentially expressed. MAML1 overexpression induced more global differential hypermethylation than hypomethylation changes. In addition, the differentially methylated regions were mapped predominantly to 3′untranslated regions, intragenic regions and gene bodies and to a lesser extent to gene regulatory sequences. Gene ontology analysis revealed that the differentially changed genes (including HOXC11, HTATIP2, SLFN12 and SOX11) are involved in the regulation of urogenital system development, cell adhesion and embryogenesis. This study is the first report that shows the global effect of a single coregulator on DNA methylation and gene expression. Our results stress and support the effects of transcriptional coregulators on the cell methylome.





Similar content being viewed by others
References
McElhinny AS, Li JL, Wu L (2008) Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene 27(38):5138–5147
Saint Just Ribeiro M, Wallberg AE (2009) Transcriptional mechanisms by the coregulator MAML1. Curr Protein Pept Sci 10(6):570–576
Hansson ML, Behmer S, Ceder R, Mohammadi S, Preta G, Grafstrom RC, Fadeel B, Wallberg AE (2012) MAML1 acts cooperatively with EGR1 to activate EGR1-regulated promoters: implications for nephrogenesis and the development of renal cancer. Plos One 7(9):e46001
Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W (2013) Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci 368(1609):20110330
Song F, Mahmood S, Ghosh S, Liang P, Smiraglia DJ, Nagase H, Held WA (2009) Tissue specific differentially methylated regions (TDMR): changes in DNA methylation during development. Genomics 93(2):130–139
Ohtani K, Dimmeler S (2011) Epigenetic regulation of cardiovascular differentiation. Cardiovasc Res 90(3):404–412
Sousa-Victor P, Munoz-Canoves P, Perdiguero E (2011) Regulation of skeletal muscle stem cells through epigenetic mechanisms. Toxicol Mech Methods 21(4):334–342
Gibney ER, Nolan CM (2010) Epigenetics and gene expression. Heredity (Edinb) 105(1):4–13
Hackett JA, Surani MA (2013) DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci 368(1609):20110328
Bergman Y, Cedar H (2013) DNA methylation dynamics in health and disease. Nat Struct Mol Biol 20(3):274–281
Hansson ML, Popko-Scibor AE, Saint Just Ribeiro M, Dancy BM, Lindberg MJ, Cole PA, Wallberg AE (2009) The transcriptional coactivator MAML1 regulates p300 autoacetylation and HAT activity. Nucleic Acids Res 37(9):2996–3006
Chen J, Li Q (2011) Life and death of transcriptional co-activator p300. Epigenetics 6(8):957–961
McKinsey TA, Zhang CL, Lu J, Olson EN (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408(6808):106–111
Blattler A, Farnham PJ (2013) Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem 288(48):34287–34294
Putnik M, Zhao C, Gustafsson JA, Dahlman-Wright K (2012) Global identification of genes regulated by estrogen signaling and demethylation in MCF-7 breast cancer cells. Biochem Biophys Res Commun 426(1):26–32
Zhao C, Putnik M, Gustafsson JA, Dahlman-Wright K (2009) Microarray analysis of altered gene expression in ERbeta-overexpressing HEK293 cells. Endocrine 36(2):224–232
da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57
da Huang W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13
Akamine R, Yamamoto T, Watanabe M, Yamazaki N, Kataoka M, Ishikawa M, Ooie T, Baba Y, Shinohara Y (2007) Usefulness of the 5′ region of the cDNA encoding acidic ribosomal phosphoprotein P0 conserved among rats, mice, and humans as a standard probe for gene expression analysis in different tissues and animal species. J Biochem Biophys Methods 70(3):481–486
Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S (2013) A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29(2):189–196
Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S (2013) ChAMP: 450 k Chip Analysis Methylation Pipeline. Bioinformatics
Wojdacz TK, Dobrovic A, Hansen LL (2008) Methylation-sensitive high-resolution melting. Nat Protoc 3(12):1903–1908
Ying Y, Tao Q (2009) Epigenetic disruption of the WNT/beta-catenin signaling pathway in human cancers. Epigenetics 4(5):307–312
Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR (2011) Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471(7336):68–73
Spitz F, Furlong EE (2012) Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13(9):613–626
Smallwood SA, Kelsey G (2012) De novo DNA methylation: a germ cell perspective. Trends Genet 28(1):33–42
Wang M, Xie H, Shrestha S, Sredni S, Morgan GA, Pachman LM (2012) Methylation alterations of WT1 and homeobox genes in inflamed muscle biopsy samples from patients with untreated juvenile dermatomyositis suggest self-renewal capacity. Arthritis Rheum 64(10):3478–3485
Shiraishi M, Sekiguchi A, Oates AJ, Terry MJ, Miyamoto Y (2002) HOX gene clusters are hotspots of de novo methylation in CpG islands of human lung adenocarcinomas. Oncogene 21(22):3659–3662
Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP (2009) Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res 11(1):R14
Lu B, Ma Y, Wu G, Tong X, Guo H, Liang A, Cong W, Liu C, Wang H, Wu M, Zhao J, Guo Y (2008) Methylation of Tip30 promoter is associated with poor prognosis in human hepatocellular carcinoma. Clin Cancer Res 14(22):7405–7412
Mavrommatis E, Fish EN, Platanias LC (2013) The schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res 33(4):206–210
Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, Muhr J (2011) Sequentially acting Sox transcription factors in neural lineage development. Genes Dev 25(23):2453–2464
Gustavsson E, Sernbo S, Andersson E, Brennan DJ, Dictor M, Jerkeman M, Borrebaeck CA, Ek S (2010) SOX11 expression correlates to promoter methylation and regulates tumor growth in hematopoietic malignancies. Mol Cancer 9:187
Wasik AM, Lord M, Wang X, Zong F, Andersson P, Kimby E, Christensson B, Karimi M, Sander B (2013) SOXC transcription factors in mantle cell lymphoma: the role of promoter methylation in SOX11 expression. Sci Rep 3:1400
Acknowledgments
This work was supported by a Grant from the Swedish Research Council to A.E.W.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2016_3946_MOESM1_ESM.pptx
Supplementary Fig. 1: Boxplots of differentially methylated probes, stratified by annotated genomic region. A. Distribution of all differentially methylated probes. B. Distribution of all differentially hypomethylated probes. C. Distribution of all differentially hypermethylated probes. The width of the boxplot reflects the number of observations in each category. UTR: untranslated region; TSS200: 200 bp within transcriptional start site; TSS1500: 1.5 kb within transcriptional start site; N shore: north shore (upstream from CpG island); N shelf: north shelf (upstream from north shore); S shore: south shore (downstream from CpG island); S shelf: south shelf (downstream from south shore). (PPTX 131 kb)
Rights and permissions
About this article
Cite this article
Putnik, M., Brodin, D., Wojdacz, T.K. et al. The transcriptional coregulator MAML1 affects DNA methylation and gene expression patterns in human embryonic kidney cells. Mol Biol Rep 43, 141–150 (2016). https://doi.org/10.1007/s11033-016-3946-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-3946-6