Abstract
Our aim was to construct infectious molecular clones of the CRF01_AE subtype in the primary infection phase of an acute HIV-1 infections in people screened from MSM populations, as well as continue preliminary research on this virus and its biological properties pertaining to deriving viruses. Walking sequencing was performed on a half-molecular clone with target fragment inserted. Western Blot was used to detect protein expression in HIV-1 infected 293T cells. Sequence analysis of HIV-1 genomic clones showed full-length HIV-1 genomic clones without frame shift mutation or termination codon. HIV-1 p24 antigens generated from 08-IMC were slightly greater than those from infectious molecular clones pNL4-3 3 and 93JP-NH1, but without statistical difference (all P > 0.05). The relative light units of 08-ISO was higher than those of 08-IMC, but no significant difference was observed (all P > 0.05). 08-IMC-driven virus was linked to lower replication kinetics. The replication levels of pNL4-3 and 08-ISO were significantly higher than the 08-IMC replication level but close to NH1 replication level (all P < 0.05). 08-IMC could infect the cells expressing CCR5 and be replicated in the CCR5-expressing cells with a positive percentage of 24.3 %, 08-ISO may use CCR5-using macrophage-tropic isolates as coreceptor, while pNL4-3 viruses with T cell tropisms utilize the CXCR4 co-receptor. Our study showed that the infectious molecular clones of viruses in the primary infection phase have a close relationship with the major prevalent CRF01_AE strains and have high homology with the viral RNA in plasma.
Similar content being viewed by others
Introduction
Human immunodeficiency virus type-1 CRF01_AE strains were first found among sex workers in Thailand in the late 1980s [1], and are prevalent among intravenous drug users (IDUs) [2]. At the end of 1994, CRF01_AE strains were first isolated in China [3], and then increased annually among sexually transmitted infections [4]. It has been reported that CRF01_AE strains have long been the predominant strain in China [5, 6]. Now available data regarding CRF01_AE is mostly derived from the studies of patients with heterosexual HIV transmission and IDUs [7–9]. In addition to other high-risk groups such as female sex workers (FSWs) and IDUs, various risk factors were detected in men who have sex with men (MSM) [10, 11].
Although we were lacking ideal animal models, we managed to assess the essential role of vitro HIV-1 infection model in the biological research on HIV-1 [12]. Namely, vitro HIV-1 infection model is a cDNA copy which contains the virus genomes in bacterial plasmids and makes cDNA itself or in vitro RNA transcription from cDNA infections [13]. Some infectious molecular clones of HIV-1, mostly from Europe, America and Africa, and only four from Thai and Japanese infected by heterosexual HIV transmission [14, 15] have been constructed [1, 16, 17]. However, they are distantly related to the current viruses CRF01_AE, and may be not beneficial for researching MSM patients with CRF01_AE infection.
Accurate description of the precise mechanism of HIV-1 transmission, along with the establishment of molecular imaging and biological processes after infection, would serve to significantly further the development of preventive measures, vaccines and microbicides [18]. Though Transmitted/Founder (T/F) strains can be obtained from blood plasma in acute infectors by SGA method, or neutralization tests using patients’ sera or neutralizing monoclonal antibodies, research on infectious molecular clones and biological characterization of these viruses have been rare in recent years [19, 20]. Currently available infectious molecular clones have only been derived from the prevalent B subtype from paid blood donors, and CRF07_BC and its subtypes have only been derived from the infected IDUs [1, 21]. There has not been any relevant report on the most prevalent CRF01_AE strains from newly infected high-risk MSM populations, let alone its subtype strains. We thus constructed the infectious molecular clones of CRF01_AE subtype strains in the primary infection phase from acute HIV-1 infectors screened from MSM populations.
Experimental materials and methods
Ethics statement
This study was conducted in complete conformity with the ethical principles in the Helsinki II Declaration. Written informed consent was obtained from all subjects before enrollment.
Study subjects
Blood specimens and early infected strains were collected from individuals of high-risk MSM groups in China with acute HIV infections of CRF01_AE subgroup strains. Viral RNA detection and HIV-1 antibodies detection were performed, the SGA method was adopted for confirming the infected T/F representative strain. Fresh PBMCs were separated using Ficoll–Hypaque density gradient centrifugation (Sigma, St. Louis, Mo.). Then, frozen stock solution was added drop wise into PBMC and stored overnight at −156 °C in a programmed cooling device.
Culture of HIV-1
The isolated PBMCs were mixed with PHA-stimulated PBMCs (HIV-), the collected supernatants (500 μl) were used to measure HIV-1 p24 antigen levels with the ELISA kit (Bio-Merieux, Marcy-l’Etoile, France) and aliquoted and stored in liquid nitrogen for later use.
Extraction of proviral DNA
A total of 200 μl of proviral DNA was extracted from 1 × 106 cultured terminal cells with the use of QIAamp DNA Blood Mini Kits (Qiagen, Hilden, Germany). Then the proviral DNA was frozen and stored at −80 °C.
Amplification of 5′ and 3′ LTR and whole genome
Nested PCR amplification was performed with the use of GoTaq PCR System (Promega). The PCR reaction volume, primers and PCR conditions are shown in Table 1. The 5′ (5′LTR-vif, nt1-5066) and 3′ (vif -3′LTR, nt4900-9719) half-molecules were amplified using a two-stage PCR. The PCR reaction volume, primers and PCR conditions.
Half-molecular cloning and extraction of plasmid DNA
The PCR products were purified and cloned using the TOPO XL PCR cloning kit (Invitrogen, Carlsbad, CA). The cloned products were transformed into One Shot cells using chemical conversion and coated on Luria–Bertani (LB) plates with kana+. Then the plates were incubated overnight at 30 °C. Single white colonies were picked from each plate and grown in 5 ml LB containing kana+. Then the colonies were proliferated at 30 °C at 200 rpm. The cloned plasmid DNA was extracted by using QIAprep Spin Miniprep kits (Qiagen). Enzyme digestion and electrophoresis were then performed.
Gene sequencing and whole-genome cloning
Walking sequencing (Huada Gene Technology Co., Ltd.) was performed on half-molecular clones with target fragment inserted. The sequencing results were edited and spliced using BioEdit software (version 5.0.6; North Carolina State University). Codon-based alignment was conducted on the spliced sequence. Double restriction enzyme digestion was performed on the 5′and 3′ half molecular clone with NdeI and NotI. The 8.5 and 5 kb fragments were collected using a Qiaquiek gel extraction kit (Qiagen. Valencia. CA) and purified and mixed together at a molar ratio of 1:2. T4 DNA ligase was then added. The half/whole-molecular clone which had no deletion or mutation or shared high sequence homology with plasma viral RNA was selected for follow-up experiments.
Transfection of plasmid DNA
The whole-genome cloned DNA was transfected into 293T cells with the use of FuGene 6 Transfection Reagent kit (Roche Diagnostics Corporation). 200 μl of supernatants were collected for the determination of HIV-1 p24 antigen, and 2 ml of supernatants were frozen and stored at −156 °C. Those with positive HIV-1 p24 antigen were selected for follow-up experiments.
Western Blot for protein expression in HIV-1 infected 293T cells
Initially, proteins were extracted from infected 293T cells referring to “Molecular Cloning”. The separated proteins in the gel were transferred to a polyvinylidene fluoride (PVDF) membrane. Goat anti-human HIV-1 were diluted in 20 ml of 10 % skim milk (1/1,000), which was then shaken for 60 min. Horseradish peroxidase-labeled goat anti-human IgG antibodies were diluted in 20 ml of 10 % skim milk (1/100,000) and incubated with PVDF membranes for 1 h. Color development was accomplished using enhanced chemiluminescence color developing kit procedures.
Identification of HIV-1 infectivity
Virus (100 μl) and TZM-b1 cell suspension (100 μl) were seeded at flat-bottomed 96 well microtiter plates (0.5 × 105 cells per well) and placed in an incubator (5 % CO2, 37 °C, 48 h). Supernatant cells (150 μl) were abandoned by adding 100 μl Bright Glo substrate solution and keeping them away from light at 37 °C for 2 min. Culture supernatant HIV-1 (150 μl) was pipetted to flat-bottomed 96 well microtiter plates with gunpoint two times. Luminometer readings were taken and recorded.
Clone-derived virus replication kinetics
The virus was prepared from 5 × 106 PBMC of HIV-1 seronegative donors by overnight adherence to plastic flasks precoated with heat-inactivated normal human serum (NHS). Nonadherent cells were removed by vigorous washing with culture media, and the virus was detached using cold Ca21- and Mg21- free PBS. Cells in triplicate wells were exposed overnight to ~400 TCID50 of virus stock and washed thoroughly. Culture supernatant HIV-1 p24 antigen was monitored three times a week during the first 21 days, and twice a week during the following 14 days. Culture supernatant from triplicate wells was pooled, and HIV-1 p24 antigen was measured in batch at the end of the assay.
HIV-1 entry coreceptor usage
HIV-1 coreceptor usage was evaluated using human HOS cells transduced with CD4, CD4 plus CCR5, or CD4 plus CXCR4, where typing was done using a modification of the GHOST cell assay. The corrected percentages of GFP-positive cells were normalized for relative cell line GFP production, reflected by parallel infections with ecotropic MLV pseudotyped HIV-1, and multiplied by the ratio.
Statistical analyses
Data was presented as mean ± standard deviation. All statistical analyses and correlation analyses in our study were conducted using the SPSS18.0 software package (SPSS Inc, Chicago, Illinois, USA). P < 0.05 was considered statistically significant.
Results
Strategy to construct the infectious molecular clones
In order to guarantee the integrity of the virus genome sequences, we developed a two-section amplification strategy, which allowed us to obtain a complete genome of HIV-1 from proviral DNA in cultured cells. Initially, PCR was amplified to obtain the 5′ and 3′ LTR at both ends of the virus. Then, based on the obtained approximate full-length gene sequences, we found the only NdeI restriction site on nt5140 gene in VIF region using NEBcutter V2.0. Finally, the obtained 5′ and 3′ 2.5 molecular clones were spliced into full-length genome clones in vitro by using the restriction enzyme site. The construction strategy is illustrated in Fig. 1.
The transfection of complete genome clone of 320008 primary isolates (08-ISO) and generation of derived virus
The human kidney cell line 293T was transfected with the genomic clones of 08-ISO. Then, after culturing it for 48 h, the culture supernatant was harvested for detection of HIV-1 p24 antigen. Each supernatant sample was distributed in two parallel wells of a 6-well plate; the virus recovered from 293T cells transfected with infectious molecular clones pNL4-3 and 93JP-NH1 were used as positive controls (Fig. 2a). The results suggested that the HIV-1 p24 antigens were detected in pNL4-3, NH1 and 08-IMC, and the quantitations of HIV-1 p24 antigen in the three groups were more than 1 × 105 pg/ml, showing evidence of the generation of the derived virus. In addition, the amount of HIV-1 p24 antigen generated from the 08-IMC was found to be slightly higher than those from infectious molecular clones pNL4-3 3 and 93JP-NH1, but without statistical difference (all P > 0.05).
a 293T cells were transfected with whole-genome cloning. PNL4-3, the subtype B of infectious molecular clone of pNL4-3; NH1,subtype CRF01_AE infectious molecular clone of 93JP-NH1; 08-IMC, 320008 whole-genome cloning; b represents identification of derivative virus infection. NH1 represents the subtype CRF01_AE of infectious molecular clone of 93JP_NH1; 08-IMC, 320008 infectious molecular clones; 08-ISO, 320008 primary viruses; RLU, the relative light units; c represents the dynamic detection of derivative virus replication. NH1 represents the subtype CRF01_AE of infectious molecular clone of 93JP_NH1; 08-IMC, 320008 infectious molecular clones; 08-ISO, 320008 primary viruses; pNL4-3, subtype B of infection molecular clone; d represents utilization of the auxiliary receptor. 08-IMC represents the 320008 infectious molecular clones; 08-ISO represents 320008 primary viruses; pNL4-3 represents subtype B infectious molecular clone; unable represents a normal cell
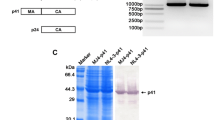
The main structural protein expression of HIV-1-drived virus
Western blot analysis of HIV-1 structural protein in the transfected 293T cells revealed that the major structural protein of the HIV-1 virus showed efficient protein expressions, such as gp160, gp120, gp41, p64, p53, p24 and p17 (Fig. 3). These results indicated that the complete genome clone constructed in the present study could be abundantly transcribed in these cell lines, and involved in the translation of the structural and functional proteins.
The infectivity of the 08-IMC-drived virus
We used the TZM-b1 indicator cells to evaluate the infectivity of the 08-IMC-drived virus to the target cells; the 08-ISO and 93JP-NH1 were taken as the positive controls. The 08-IMC-drived virus, 08-ISO and 93JP-NH1 (per 1 ng) were seeded simultaneously in three parallel wells of a 6-well culture plate containing TZM-b1cells. The results showed that the three viruses were able to infect TZM-b1 cells to with different degrees. The relative light unit of the 08-ISO was higher than those in 08-IMC which shared a similar relative light unit with NH1, but no significant difference was observed (Fig. 2b).
The replication kinetics of 08-IMC-drived virus
CD8-PBMC were isolated from the PBMC and used as host cells in the infection test enhanced by the use of DEAE. The pNL4-3, NH1 and 08-ISO were taken as control groups. The cell-culture supernatants were captured once every two days to draw the viral replication kinetics curve (Fig. 2c). We found that 08-IMC-drived viruses were linked to lower replication kinetics than 08-ISO; the replication levels of pNL4-3 and 08-ISO were significantly higher than the 08-IMC replication level which was close to NH1 replication level (all P < 0.05). In addition, p24 expression decreased rapidly 10 days after infection.
The 08-IMC-drived virus in the context of coreceptor usage
GHOST-CD4-CXCR4 and GHOST-CD4-CCR5 cells were infected by each virus, fixed and analyzed with flow cytometry. Our study revealed that 08-IMC could infect the cells expressing CCR5 and be replicated in the CCR5-expressing cells with a positive percentage of 24.3 %, suggesting that 08-IMC used CCR5 as the coreceptor with macrophage tropism. Additionally, our results indicated that 08-ISO was determined to use CCR5-using macrophage-tropic isolates (R5 viruses) as coreceptor; while pNL4-3 viruses with T-cell tropism utilize the CXCR4 co-receptor (Fig. 2d).
Discussion
The research presented here was conducted on CRF01_AE subtype virus in primary infection phase from an acute HIV-1 infector in MSM populations. We constructed infectious molecular clones of viruses in primary infection phase and investigated their biological characterization. We observed that the 08-IMC and NH1 had packed more viruses after being transfected with 293T cells, but the infection and replication ability were significantly lower than those of pNL4-3. These results suggested that pNL4-3 may have packed more viruses that can establish effective viral infections. By analyzing and comparing these three infectious molecular clones, we found that pNL4-3 may be a chimeric virus composed of the 5′ end of NY 5 and 3′ terminal fragment of LAV, which had higher activation of infection and replication [22, 23]. Furthermore, 08-IMC and NH1 were derived from acute CRF01_AE infection and chronic CRF01_AE infection, respectively, while the pNL4-3 was separated from AIDS subtype B infection. Viruses in different course of disease have quite different infection and replication ability, and the subtype B and C T/F strains showed a higher replication ability in PBMC [24]. Hence, we hypothesized that their replication ability might be associated with the virus subtypes. However, replication ability of infectious molecular clones has also been revealed to be limited in PBMC, while over-expressed in CD4 cell lines [25]. The possible reason for this is that the infectious molecular clones reproduced in the target cells via over-expressed CCR5 or CD4. Or, the infectious molecular clones may occur because some viruses slowly reproduce in PBMC. Therefore, to improve a series of clones in terms of infection and replication ability for the sake of vaccine development, further genetic modification to env region or combination with other genes should be considered. In addition, an inevitable introduction of a few point mutations and exogenous sequences in the construction process has possibly resulted in lower virulence of derived viruses than the wild-types.
It has been reported that CCR5 HIV-1 strains were in vivo in persons with early HIV infection [26]. With progression of the disease, three different HIV-1 strains, including CCR5, CXCR4/CCR5 and CXCR4, can coexist in vivo in some HIV-infected persons [27]. At the terminal stage of infection, CCR5 HIV-1 strains in 50 % of the HIV-infected persons would transform into CXCR4 HIV-1 strains [28]. The primary human immunodeficiency virus type 1 isolates and infectious molecular clones of HIV that we obtained showed CCR5 tropism without any changes, which were affected by integration host factor, in vitro culture and other factors [29]. Compared with dual-tropic NH1 cloning, this CCR5 strain has unique superiority, and can well simulate the model of CRF01_AE strains in individuals who were infected naturally [30]. Moreover, the infectious molecular clone of CCR5 tropism that we constructed may play crucial roles in the evaluation of potential HIV-1 vaccines [31, 32]. The infectious molecular clone of CCR5 tropism can provide a basis for better understanding the biological characteristics of HIV-1 isolates, and provide assistance to the researches on hosts’ cellular immune responses [1, 32]. In addition, it can also determine the specific cytotoxic T lymphocyte (CTL) epitope [33].
In conclusion, our study indicated that the infectious molecular clones of viruses in primary infection phase had a close relationship with the most prevalent CRF01_AE strains and had high homology with the viral RNA in plasma, which may play a pivotal role in future research on the HIV-1 CRF01_AE subtype, especially the biological characterization of the prevalent CRF01_AE subtype strains in China MSM populations. Finally, it could be crucial for drug screening and for evaluating vaccine candidates.
References
Wang Z, Hong K, Zhang J, Zhang L, Li D, Ren L, Liang H, Shao Y (2013) Construction and characterization of highly infectious full-length molecular clones of a HIV-1 CRF07_BC isolate from Xinjiang, China. PLoS One 8:e79177. doi:10.1371/journal.pone.0079177
Kijak GH, Beyrer C, Tovanabutra S, Sripaipan T, Suriyanon V, Moqueet N, Sanders-Buell E, Saokhieo P et al (2011) Socio-demographic and drug use factors associated with HIV-1 recombinants and dual infections in Northern Thai drug users: associations of risk with genetic complexity. Drug Alcohol Depend 116:24–30. doi:10.1016/j.drugalcdep.2010.11.013
Feng Y, He X, Hsi JH, Li F, Li X, Wang Q, Ruan Y, Xing H et al (2013) The rapidly expanding CRF01_AE epidemic in China is driven by multiple lineages of HIV-1 viruses introduced in the 1990s. AIDS 27:1793–1802. doi:10.1097/QAD.0b013e328360db2d
Ye J, Xin R, Yu S, Bai L, Wang W, Wu T, Su X, Lu H et al (2013) Phylogenetic and temporal dynamics of human immunodeficiency virus type 1 CRF01_AE in China. PLoS One 8:e54238. doi:10.1371/journal.pone.0054238
Zeng H, Sun Z, Liang S, Li L, Jiang Y, Liu W, Sun B, Li J et al (2012) Emergence of a new HIV type 1 CRF01_AE variant in Guangxi, Southern China. AIDS Res Hum Retroviruses 28:1352–1356. doi:10.1089/AID.2011.0364
An M, Han X, Xu J, Chu Z, Jia M, Wu H, Lu L, Takebe Y et al (2012) Reconstituting the epidemic history of HIV strain CRF01_AE among men who have sex with men (MSM) in Liaoning, northeastern China: implications for the expanding epidemic among MSM in China. J Virol 86:12402–12406. doi:10.1128/JVI.00262-12
Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P et al (2008) Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol 82:3952–3970. doi:10.1128/JVI.02660-07
Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T et al (2008) Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA 105:7552–7557. doi:10.1073/pnas.0802203105
Keele BF (2010) Identifying and characterizing recently transmitted viruses. Curr Opin HIV AIDS 5:327–334. doi:10.1097/COH.0b013e32833a0b9b
Grant RM, Lama JR, Anderson PL, McMahan V, Liu AY, Vargas L, Goicochea P, Casapia M et al (2010) Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med 363:2587–2599. doi:10.1056/NEJMoa1011205
Finlayson TJ, Le B, Smith A, Bowles K, Cribbin M, Miles I, Oster AM, Martin T et al. (2011) HIV risk, prevention, and testing behaviors among men who have sex with men–National HIV Behavioral Surveillance System, 21 U.S. cities, United States, 2008. MMWR Surveill Summ 60: 1–34. http://www.ncbi.nlm.nih.gov/pubmed/?term=HIV+risk%2C+prevention%2C+and+418+testing+behaviors+among+men+who+have+sex+with+men%C3%90National+419+HIV+Behavioral+Surveillance+System
Haase AT (2010) Targeting early infection to prevent HIV-1 mucosal transmission. Nature 464:217–223. doi:10.1038/nature08757
Bahuon C, Despres P, Pardigon N, Panthier JJ, Cordonnier N, Lowenski S, Richardson J, Zientara S et al (2012) IS-98-ST1 West Nile virus derived from an infectious cDNA clone retains neuroinvasiveness and neurovirulence properties of the original virus. PLoS One 7:e47666. doi:10.1371/journal.pone.0047666
Zhang Y, Lu L, Ba L, Liu L, Yang L, Jia M, Wang H, Fang Q et al (2006) Dominance of HIV-1 subtype CRF01_AE in sexually acquired cases leads to a new epidemic in Yunnan province of China. PLoS Med 3:e443. doi:10.1371/journal.pmed.0030443
Hemelaar J, Gouws E, Ghys PD, Osmanov S, Isolation W-UNfH, Characterisation (2011) Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS 25:679–689. doi:10.1097/QAD.0b013e328342ff93
Tee KK, Kusagawa S, Li XJ, Onogi N, Isogai M, Hase S, Uenishi R, Liao H et al (2009) Isolation and characterization of a replication-competent molecular clone of an HIV-1 circulating recombinant form (CRF33_01B). PLoS One 4:e6666. doi:10.1371/journal.pone.0006666
Baalwa J, Wang S, Parrish NF, Decker JM, Keele BF, Learn GH, Yue L, Ruzagira E et al (2013) Molecular identification, cloning and characterization of transmitted/founder HIV-1 subtype A, D and A/D infectious molecular clones. Virology 436:33–48. doi:10.1016/j.virol.2012.10.009
Kondo M, Lemey P, Sano T, Itoda I, Yoshimura Y, Sagara H, Tachikawa N, Yamanaka K et al (2013) Emergence in Japan of an HIV-1 variant associated with transmission among men who have sex with men (MSM) in China: first indication of the International Dissemination of the Chinese MSM lineage. J Virol 87:5351–5361. doi:10.1128/JVI.02370-12
Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R et al (2012) Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol 86:2715–2728. doi:10.1128/JVI.06157-11
Wilen CB, Parrish NF, Pfaff JM, Decker JM, Henning EA, Haim H, Petersen JE, Wojcechowskyj JA et al (2011) Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J Virol 85:8514–8527. doi:10.1128/JVI.00736-11
Zhang Q, Zhang X, Wu H, Seto D, Zhang HJ, Chen Z, Wan C, Zheng BJ (2012) Parental LTRs are important in a construct of a stable and efficient replication-competent infectious molecular clone of HIV-1 CRF08_BC. PLoS One 7:e31233. doi:10.1371/journal.pone.0031233
Chhatwal P, Bankwitz D, Gentzsch J, Frentzen A, Schult P, Lohmann V, Pietschmann T (2012) Bile acids specifically increase hepatitis C virus RNA-replication. PLoS One 7:e36029. doi:10.1371/journal.pone.0036029
Troyer RM, Thompson J, Elder JH, VandeWoude S (2013) Accessory genes confer a high replication rate to virulent feline immunodeficiency virus. J Virol 87:7940–7951. doi:10.1128/JVI.00752-13
Lau KA, Wang B, Saksena NK (2007) Emerging trends of HIV epidemiology in Asia. AIDS Rev 9: 218–229. http://www.ncbi.nlm.nih.gov/pubmed/18219365
Rodriguez MA, Chen Y, Craigo JK, Chatterjee R, Ratner D, Tatsumi M, Roy P, Neogi D et al (2006) Construction and characterization of an infectious molecular clone of HIV-1 subtype A of Indian origin. Virology 345:328–336. doi:10.1016/j.virol.2005.09.053
Zhang Y, de Lara C, Worth A, Hegedus A, Laamanen K, Beverley P, Macallan D (2013) Accelerated in vivo proliferation of memory phenotype CD4+ T-cells in human HIV-1 infection irrespective of viral chemokine co-receptor tropism. PLoS Pathog 9:e1003310. doi:10.1371/journal.ppat.1003310
Bon I, Lembo D, Rusnati M, Clo A, Morini S, Miserocchi A, Bugatti A, Grigolon S et al (2013) Peptide-derivatized SB105-A10 dendrimer inhibits the infectivity of R5 and X4 HIV-1 strains in primary PBMCs and cervicovaginal histocultures. PLoS One 8:e76482. doi:10.1371/journal.pone.0076482
Nishimura Y, Shingai M, Lee WR, Sadjadpour R, Donau OK, Willey R, Brenchley JM, Iyengar R et al (2011) Recombination-mediated changes in coreceptor usage confer an augmented pathogenic phenotype in a nonhuman primate model of HIV-1-induced AIDS. J Virol 85:10617–10626. doi:10.1128/JVI.05010-11
Ping LH, Joseph SB, Anderson JA, Abrahams MR, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L et al (2013) Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol 87:7218–7233. doi:10.1128/JVI.03577-12
Panos G, Watson DC (2014) Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition. Crit Rev Microbiol. doi:10.3109/1040841X.2013.867829
Saunders KO, Rudicell RS, Nabel GJ (2012) The design and evaluation of HIV-1 vaccines. AIDS 26:1293–1302. doi:10.1097/QAD.0b013e32835474d2
McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF (2010) The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 10:11–23. doi:10.1038/nri2674
Nomura T, Matano T (2012) Association of MHC-I genotypes with disease progression in HIV/SIV infections. Front Microbiol 3:234. doi:10.3389/fmicb.2012.00234
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper.
Competing interests
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Additional information
The Publisher and Editor retract this article in accordance with the recommendations of the Committee on Publication Ethics (COPE). After a thorough investigation we have strong reason to believe that the peer review process was compromised.
About this article
Cite this article
Wang, HW., Zhu, B., Hou, LJ. et al. RETRACTED ARTICLE: An infectious molecular clone in early infection with HIV-1 subtype CRF01_AE strains: construction and biological properties. Mol Biol Rep 42, 329–336 (2015). https://doi.org/10.1007/s11033-014-3754-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3754-9