Abstract
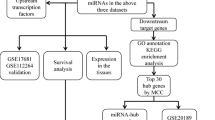
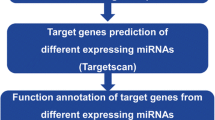
Lung cancer is a worldwide leading cause of cancer-related death. The aim of this study was to identify target genes and specific biomarkers for identification and treatment of different types of lung cancer with DNA microarray. Gene expression profile GSE6044 and miRNA microarray profile GSE17681 were downloaded from Gene Expression Omnibus database. The differentially expressed genes (DEGs) and miRNAs were screened with multtest package in R language. Then, functional enrichment analysis of identified DEGs was performed. Furthermore, the verified target genes based on screened miRNAs were selected from miRTarBase and miRecords databases. Then miRNA-target gene regulation network was constructed. APOE, CDC6 and ATP2B1were involved in most of the functions obtained for adenocarcinomas, small cell lung cancer and squamous cell carcinomas, respectively. The target DEGs of differentially expressed hsa-miR-29a included FGG in adenocarcinoma, RAN and COL4A1 in small cell lung cancer, GLUL in squamous cell carcinoma. The target DEGs of has-miR-7 were SNCA and SLC7A5 in adenocarcinoma and small cell lung cancer, respectively. ICAM1 and KIT were the target DEGs of hsa-miR-222 in adenocarcinoma and squamous cell carcinoma. The miRNAs and their differentially expressed target genes have the potential to be used in clinic for diagnosis and treatment of different kinds of lung cancer in the future.



Similar content being viewed by others
References
Boyle P, Chapman C, Holdenrieder S, Murray A, Robertson C, Wood W, Maddison P, Healey G, Fairley G, Barnes A (2011) Clinical validation of an autoantibody test for lung cancer. Ann Oncol 22(2):383–389
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90
Ettinger DS, Akerley W, Bepler G, Blum MG, Chang A, Cheney RT, Chirieac LR, D’Amico TA, Demmy TL, Ganti AKP (2010) Non–small cell lung cancer. J Natl Compr Canc Netw 8(7):740–801
Ebert W, Muley T, Drings P (1996) Does the assessment of serum markers in patients with lung cancer aid in the clinical decision making process? Anticancer Res 16(4B):2161–2168
Ando S, Suzuki M, Yamamoto N, Iida T, Kimura H (2004) The prognostic value of both neuron-specific enolase (NSE) and Cyfra21-1 in small cell lung cancer. Anticancer Res 24(3B):1941–1946
Mulshine JL, Sullivan DC (2005) Lung cancer screening. N Engl J Med 352(26):2714–2720
Pastorino U (2010) Lung cancer screening. Br J Cancer 102(12):1681–1686
Brennan P, Hainaut P, Boffetta P (2011) Genetics of lung-cancer susceptibility. Lancet Oncol 12(4):399–408. doi:10.1016/S1470-2045(10)70126-1
Wang L, Xiong Y, Sun Y, Fang Z, Li L, Ji H, Shi T (2010) HLungDB: an integrated database of human lung cancer research. Nucleic Acids Res 38(Suppl 1):D665–D669
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C (2010) Serum MicroRNA signatures identified in a genome-wide serum MicroRNA expression profiling predict survival of non–small-cell lung cancer. J Clin Oncol 28(10):1721–1726
Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V (2010) TGF-β IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci 107(35):15535–15540
Spies M, Dasu MR, Svrakic N, Nesic O, Barrow RE, Perez-Polo JR, Herndon DN (2002) Gene expression analysis in burn wounds of rats. Am J Physiol Regul Integr Comp Physiol 283(4):R918–R930
Eb WDT, Muller-Hermelink H (2004) World Health Organization classification of tumors: Pathology and genetics of tumors of the lung, pleura, thymus and heart. IARC Press, Lyon
Rohrbeck A, Neukirchen J, Rosskopf M, Pardillos GG, Geddert H, Schwalen A, Gabbert HE, von Haeseler A, Pitschke G, Schott M (2008) Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J Transl Med 6:69
Keller A, Leidinger P, Borries A, Wendschlag A, Wucherpfennig F, Scheffler M, Huwer H, Lenhof H-P, Meese E (2009) miRNAs in lung cancer-studying complex fingerprints in patient’s blood cells by microarray experiments. BMC Cancer 9(1):353
Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB (2001) Missing value estimation methods for DNA microarrays. Bioinformatics 17(6):520–525
André F, João S, Leonardo R, Carlos F (2006) Evaluating different methods of microarray data normalization. BMC Bioinformatics 7(1):469
Dudoit S, Shaffer JP, Boldrick JC (2003) Multiple hypothesis testing in microarray experiments. Stat Sci 18(1):71–103
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 57(1):289–300
Lewis CI, von Leibniz GWF (1918) A survey of symbolic logic. University of California Press, Berkeley, CA
Nam D, Kim S-Y (2008) Gene-set approach for expression pattern analysis. Brief Bioinform 9(3):189–197
Allison DB, Cui X, Page GP, Sabripour M (2006) Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7(1):55–65
Da Wei Huang BTS, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57
Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T (2009) miRecords: an integrated resource for microRNA–target interactions. Nucleic Acids Res 37(Suppl 1):D105–D110
Hsu S-D, Lin F-M, Wu W-Y, Liang C, Huang W-C, Chan W-L, Tsai W-T, Chen G-Z, Lee C-J, Chiu C-M (2011) miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Res 39(Suppl 1):D163–D169
Parkin DM, Bray F, Devesa S (2001) Cancer burden in the year 2000. The global picture. Eur J Cancer 37:4–66
Pirozynski M (2009) 100 years of lung cancer (Retraction of vol 100, pg 2073, 2006). Respir Med 103(8):1244
Wang Y, Chen Z, Chen J, Pan J, Zhang W, Pan Q, Ding H, Lin X, Wen X, Li Y (2013) The diagnostic value of apolipoprotein E in malignant pleural effusion associated with non-small cell lung cancer. Clin Chim Acta 421:230–235
Su W-P, Chen Y-T, Lai W-W, Lin C-C, Yan J-J, Su W-C (2011) Apolipoprotein E expression promotes lung adenocarcinoma proliferation and migration and as a potential survival marker in lung cancer. Lung Cancer 71(1):28–33
Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters J-M, Helin K (2000) Cell cycle–and cell growth–regulated proteolysis of mammalian CDC6 is dependent on APC–CDH1. Genes Dev 14(18):2330–2343
Basaki Y, K-i Taguchi, Izumi H, Murakami Y, Kubo T, Hosoi F, Watari K, Nakano K, Kawaguchi H, Ohno S, Kohno K, Ono M, Kuwano M (2010) Y-box binding protein-1 (YB-1) promotes cell cycle progression through CDC6-dependent pathway in human cancer cells. Eur J Cancer 46(5):954–965. doi:10.1016/j.ejca.2009.12.024
Kobayashi Y, Hirawa N, Tabara Y, Muraoka H, Fujita M, Miyazaki N, Fujiwara A, Ichikawa Y, Yamamoto Y, Ichihara N, Saka S, Wakui H, Yoshida S-I, Yatsu K, Toya Y, Yasuda G, Kohara K, Kita Y, Takei K, Goshima Y, Ishikawa Y, Ueshima H, Miki T, Umemura S (2012) Mice lacking hypertension candidate Gene ATP2B1 in vascular smooth muscle cells show significant blood pressure elevation. Hypertension 59(4):854–860. doi:10.1161/hypertensionaha.110.165068
Monteith GR, McAndrew D, Faddy HM, Roberts-Thomson SJ (2007) Calcium and cancer: targeting Ca2+ transport. Nat Rev Cancer 7(7):519–530
Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clinical Lung Cancer 10(1):42–46. doi:10.3816/CLC.2009.n.006
Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM (2010) Effects of MicroRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 51(3):836–845
Fort A, Borel C, Migliavacca E, Antonarakis SE, Fish RJ, Neerman-Arbez M (2010) Regulation of fibrinogen production by microRNAs. Blood 116(14):2608–2615
Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lü J (2011) miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 45(2):287–294
Plaisier CL, Pan M, Baliga NS (2012) A miRNA-regulatory network explains how dysregulated miRNAs perturb oncogenic processes across diverse cancers. Genome Res 22(11):2302–2314
Gilad S, Lithwick-Yanai G, Barshack I, Benjamin S, Krivitsky I, Edmonston TB, Bibbo M, Thurm C, Horowitz L, Huang Y, Feinmesser M, Steve Hou J, St Cyr B, Burnstein I, Gibori H, Dromi N, Sanden M, Kushnir M, Aharonov R (2012) Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J Mol Diagn 14(5):510–517. doi:10.1016/j.jmoldx.2012.03.004
Kaira K, Oriuchi N, Imai H, Shimizu K, Yanagitani N, Sunaga N, Hisada T, Tanaka S, Ishizuka T, Kanai Y (2008) Prognostic significance of L-type amino acid transporter 1 expression in resectable stage I-III nonsmall cell lung cancer. Br J Cancer 98(4):742–748
Kido Y, Tamai I, Uchino H, Suzuki F, Sai Y, Tsuji A (2001) Molecular and functional identification of large neutral amino acid transporters LAT1 and LAT2 and their pharmacological relevance at the blood-brain barrier. J Pharm Pharmacol 53(4):497–503
Miko E, Margitai Z, Czimmerer Z, Várkonyi I, Dezső B, Lányi Á, Bacsó Z, Scholtz B (2011) miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett 585(8):1191–1196
Arnold CP, Tan R, Zhou B, Yue S-B, Schaffert S, Biggs JR, Doyonnas R, Lo M-C, Perry JM, Renault VM (2011) MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res 21(5):798–810
Lin Q, Mao W, Shu Y, Lin F, Liu S, Shen H, Gao W, Li S, Shen D (2012) A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non-small-cell lung cancer by stem-loop RT-PCR. J Cancer Res Clin Oncol 138(1):85–93
Guo L, Ji X, Yang S, Hou Z, Luo C, Fan J, Ni C, Chen F (2013) Genome-wide analysis of aberrantly expressed circulating miRNAs in patients with coal workers’ pneumoconiosis. Mol Biol Rep 40(5):3739–3747. doi:10.1007/s11033-012-2450-x
Thanopoulou E, Kotzamanis G, Pateras I, Ziras N, Papalambros A, Mariolis-Sapsakos T, Sigala F, Johnson E, Kotsinas A, Scorilas A, Gorgoulis V (2012) The single nucleotide polymorphism g.1548A > G (K469E) of the ICAM-1 gene is associated with worse prognosis in non-small cell lung cancer. Tumor Biol 33(5):1429–1436. doi:10.1007/s13277-012-0393-4
Guney N, Soydinc HO, Derin D, Tas F, Camlica H, Duranyildiz D, Yasasever V, Topuz E (2008) Serum levels of intercellular adhesion molecule ICAM-1 and E-selectin in advanced stage non-small cell lung cancer. Med Oncol 25(2):194–200
Blagosklonny MV, Pardee AB (2001) Exploiting cancer cell cycling for selective protection of normal cells. Cancer Res 61(11):4301–4305
Acknowledgments
We wish to express our warm thanks to all the authors who contributed a lot to the research.
Author information
Authors and Affiliations
Corresponding authors
Additional information
The Publisher and Editor retract this article in accordance with the recommendations of the Committee on Publication Ethics (COPE). After a thorough investigation we have strong reason to believe that the peer review process was compromised.
About this article
Cite this article
Zhou, C., Chen, H., Han, L. et al. RETRACTED ARTICLE: Identification of featured biomarkers in different types of lung cancer with DNA microarray. Mol Biol Rep 41, 6357–6363 (2014). https://doi.org/10.1007/s11033-014-3515-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3515-9