Abstract
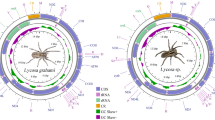
In this study, the complete mitochondrial genome of the Eurasian flying squirrel Pteromys volans (Rodentia, Sciuromorpha, Sciuridae) was sequenced and characterized in detail. The entire mitochondrial genome of P. volans consisted of 16,513 bp and contained 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and two non-coding regions. Its gene arrangement pattern was consistent with the mammalian ground pattern. The overall base composition and AT contents were similar to those of other rodent mitochondrial genomes. The light-strand origin generally identified between tRNA Asn and tRNA Cys consisted of a secondary structure with an 11-bp stem and an 11-bp loop. The large control region was constructed of three characteristic domains, ETAS, CD, and CSB without any repeat sequences. Each domain contained ETAS1, subsequences A, B, and C, and CSB1, respectively. In order to examine phylogenetic contentious issues of the monophyly of rodents and phylogenetic relationships among five rodent suborders, here, phylogenetic analyses based on nucleotide sequence data of the 35 rodent and 3 lagomorph mitochondrial genomes were performed using the Bayesian inference and maximum likelihood method. The result strongly supported the rodent monophyly with high node confidence values (BP 100 % in ML and BPP 1.00 in BI) and also monophylies of four rodent suborders (BP 85–100 % in ML and BPP 1.00 in BI), except for Anomalumorpha in which only one species was examined here. Also, phylogenetic relationships among the five rodent suborders were suggested and discussed in detail.



Similar content being viewed by others
References
Cobet GB, Hill JE (1991) A world list of mammalian species, 3rd edn. Oxford University Press, Oxford
Nowak RM (1999) Walker’s mammals of the world, 6th edn. Johns Hopkins University Press, Baltimore
Lee MY, Park SK, Hong YJ, Kim YJ, Voloshina I, Myslenkov A, Saveljev AP, Choi TY, Piao RZ, An JH, Lee MH, Lee H, Min MS (2008) Mitochondrial genetic diversity and phylogenetic relationship of Siberian flying squirrel (Pteromys volans). Anim Cell Syst 12:269–277
Won C, Smith KG (1999) History and current status of mammals of the Korea Peninsula. Mamm Rev 29:3–33
Wilson DE, Reeder DM (2005) Mammal species of the world. A taxonomic and geographic reference 3rd ed. Johns Hopkins University Press, Baltimore
Hartenberger JL (1985) The order Rodentia: major questions on their evolutionary origin, relationships and suprafamilial systematics. In: Luckett WP, Hartenberger JL (eds) Evolutionary relationships among rodents: a multidisciplinary analysis. Plenum Press, New York, pp 1–33
Luckett WP, Hartenberger JL (1993) Monophyly or polyphyly of rodentia: possible conflict between morphological and molecular interpretations. J Mamm Evol 1:127–147
Lin YH, Waddell P, Penny D (2002) Pika and vole mitochondrial genomes increase support for both rodent monophyly and Glires. Gene 294:119–129
Douzery EJP, Huchon D (2004) Rabbits, if anything, are likely Glires. Mol Phylogenet Evol 33:922–935
Horner DS, Lefkimmiatis K, Reyes A, Gissi C, Saccone C, Pesole G (2007) Phylogenetic analyses of complete mitochondrial genome sequences suggest a basal divergence of the enigmatic rodent Anomalurus. BMC Evol Biol 8:7–16
Blanga-Kanfi S, Miranda H, Penn O, Pupko T, Debry RW, Huchon D (2009) Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol Biol 9:71
Graur D, Hide WA, Li WH (1991) Is the guinea pig a rodents? Nature 351:649–652
D’Erchia AM, Gissi C, Pesole G, Saccone C, Arnason U (1996) The guinea pig is not rodent. Nature 381:597–600
Graur D, Duret L, Gouy M (1996) Phylogenetic position of the order Lagomorpha (rabbits, hares and allies). Nature 376:333–335
Reyes A, Pesole G, Saccone C (1998) Complete mitochondrial DNA sequence of the fat dormouse, Glis glis: further evidence of rodent paraphyly. Mol Biol Evol 15:499–505
Reyes A, Gissi C, Pesole G, Catzeflis FM, Saccone C (2000) Where do rodents fit? Evidence from the complete mitochondrial genome of Sciurus vulgaris. Mol Biol Evol 17:979–983
Brant JF (1855) Beiträge zur nahern Kenntniss der Säugethiere Russlands. Mem Acad Imp St Petersbourg Ser 69:1–375
Tullberg T (1899) Ueber das System Der Nagetiere: Ein Phylogenetische Studie. Nuova Acta Reg Soc Sic Upsala Ser 3:1–514
Nedbal MA, Honeycutt RL, Schlitter DA (1996) Higher-level systematics of rodents (Mammalia, Rodentia): evidence from the mitochondrial 12S rRNA gene. J Mamm Evol 3:201–237
Montgelard C, Bentz S, Tirard C, Verneau O, Catzeflis FM (2002) Molecular systematics of Sciurognathi (Rodentia): the mitochondrial cytochrome b and 12S rRNA genes support the Anomaluroidea (Pedetidae and Anomaluridae). Mol Phylogenet Evol 22:220–233
Huchon D, Madsen O, Sibbald MJJB, Ament K, Stanhope M, Catzeflis F, De Jong WW, Douszeyr EJP (2002) Rodent phylogeny and a timescale for the evolution of glires: evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol 19(7):1053–1065
DeBry RW (2003) Identifying conflicting signal in a multigene analysis reveals a highly resolved tree: the phylogeny of Rodentia (Mammalia). Syst Biol 52(5):604–617
Carleton MD, Musser GG (2005) In: Wilson DE, Reeder DM, Baltimore MD (eds) Mammal species of the world; a taxonomic and geographic reference Volume 2, 3rd edn. Johns Hopkins University press, Baltimore, pp 745–751
Montgelard C, Forty E, Arnal V, Matthee CA (2008) Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol Biol 8:321
Horn S, Durka W, Wolf R, Ermala A, Stubbe A, Stubbe M, Hofreiter M (2011) Mitochondrial genomes reveal slow rates of molecular evolution and the timing of speciation in beavers (Castor), one of the largest rodent species. PLoS One 6(1):e14622
Lim JT, Hwang UW (2006) The complete mitochondrial genome of Pollicipes mitella (Crustacea, Maxillopoda, Cirripedia): non-monophylies of Maxillipoda and Crustacea. Mol Cells 22:314–322
Choi EH, Park SJ, Jang KH, Hwang UW (2007) Complete mitochondrial genome of a Chinese scorpion Mesobuthus martensii (Chelicerata, Scorpiones, Buthidae). DNA Seq 18:461–473
Ryu SH, Lee JM, Jang KH, Choi EH, Park SJ, Chang CY, Kim W, Hwang UW (2007) Partial mitochondrial gene arrangements support a close relationship between Tardigrada and Arthropoda. Mol Cells 24(3):351–375
Jang KH, Hwang UW (2009) Complete mitochondrial genome of Bugula neritina (Bryozoa, Gymnolaemata, Cheilostomata): phylogenetic position of Bryozoa and phylogeny of lophophorates within the Lophotrochozoa. BMC Genomics 21:167–285
Jang KH, Ryu SH, Hwang UW (2009) Mitochondrial genome of the Eurasian otter Lutra lutra (Mammalia, Carnivora, Mustelidae). Gene Genomics 31:19–27
Choi EH, Kim SK, Ryu SH, Jang KH, Hwang UW (2010) Mitochondrial genome phylogeny among Asiatic black bear Ursus thibetanus subspecies and comprehensive analysis of their control regions. Mitochondrial DNA 21:105–114
Ryu SH, Hwang UW (2011) Complete mitochondrial genome of the Seoul frog Rana chosenica (Amphibia, Ranidae): comparison of R. chosenica and R. plancyi. Mitochondrial DNA 22(3):53–54
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp Ser 41:95–98
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X window interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res 24:4876–4882
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acid Res 25:955–964
Larizza A, Pesole G, Reyes A, Sbisà E, Saccone C (2002) Lineage specificity of the evolutionary dynamics of the mtDNA D-loop region in rodents. J Mol Evol 54:754–755
Catanesi CI, Vidal-Rioja L, Zambelli A (2006) Molecular and phylogenetic analysis of mitochondrial control region in robertsonian karyomorphs of Graomys griseoflavus (Rodentia, Signodontinae). Mastozoología Neotropical 13(1):21–30
Tomasco IH, Lessa EP (2011) The evolution of mitochondrial genomes in subterranean caviomorph rodents: adaptation against a background of purifying selection. Mol Phylogenet Evol 61(1):64–70
Mouchaty S, Catzeflis F, Janke A, Arnason U (2001) Molecular evidence of an African Phiomorpha-South American Caviomorpha clade and support for Hystricognathi based on the complete mitochondrial genome of the cane rat (Thryonomys swinderianus). Mol Phylogenet Evol 18:127–135
Partridge MA, Davidson MM, Hei TK (2007) The complete nucleotide sequence of Chinese hamster (Cricetulus griseus) mitochondrial DNA. DNA Seq 18(5):341–346
Triant DA, Dewoody JA (2006) Accelerated molecular evolution in Microtus (Rodentia) as assessed via complete mitochondrial genome sequences. Genetica 128(1–3):95–108
Fan L, Fan Z, Yue H, Zhang X, Liu Y, Sun Z, Liu S, Yue B (2011) Complete mitochondrial genome sequence of the Chinese scrub vole (Neodon irene). Mitochondrial DNA 22(3):50–52
Nilsson MA, Härlid A, Kullberg M, Janke A (2010) The impact of fossil calibrations, codon positions and relaxed clocks on the divergence time estimates of the native Australian rodents (Conilurini). Gene 55(1–2):22–31
Gregorová S, Divina P, Storchova R, Trachtulec Z, Fotopulosova V, Svenson KL, Donahue LR, Paigen B, Forejt J (2008) Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res 18(3):509–515
Robins JH, McLenachan PA, Phillips MJ, Craig L, Ross HA, Matisoo-Smith E (2008) Dating of divergences within the Rattus genus phylogeny using whole mitochondrial genomes. Mol Phylogenet Evol 49(2):460–466
Arnason U, Adegoke JA, Bodin K, Born EW, Esa YB, Gullberg A, Nilsson M, Short RV, Xu X, Janke A (2002) Mammalian mitogenomic relationships and the root of the eutherian tree. Proc Natl Acad Sci 99(12):8151–8156
Gissi C, Gullberg A, Arnason U (1998) The complete mitochondrial DNA sequence of the rabbit, Oryctolagus cuniculus. Genomics 50(2):161–169
Castresana J (2000) Selection of conserved block from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Posada D, Crandall K (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Lartillot N, Lepage T, Blanquart S (2009) PhyloBayes 3. A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25(17):2286–2288
Garey JR, Wolstenholme DR (1989) Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNAser (AGN) that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol 28:374–387
Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474
Hixson JE, Wong TN, Clayton DA (1986) Both the conserved stem-loop and divergent 5′-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J Biol Chem 261:2384–2390
Sbisà E, Tanzariello F, Reyes A, Pesole G, Saccone C (1997) Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and their functional and evolutionary implications. Gene 31:125–140
Jackson DA, Bartlett J, Cook PR (1996) Sequences attaching loops of nuclear and mitochondrial DNA to underlying structures in human cells: the role of transcription units. Nucleic Acid Res 24:1212–1219
Carrol RL (1988) Vertebrate paleontology and evolution. W. H. Freeman, New York
Saveur G, Macaya G, Kadi F, Bernardi G (1993) The isochore patterns of mammalian genomes and their phylogenetic implications. J Mol Evol 37:93–108
Lavocat R, Parent JP (1985) Phylogenetic analyses of middle ear features in fossil and living rodents. In: Luckett WP, Hartenberger JL (eds) Evolutionary relationships among rodents: a multidisciplinary analysis. Plenum Press, New York, pp 333–354
Sannier J, Gerbault-Seureau M, Dutrillaux B, Richard FA (2011) Conserved although very different karyotypes in Gliridae and Sciuridae and their contribution to chromosomal signatures in Glires. Cytogenet Genome Res 134:51–63
Adkins RM, Walton AH, Honeycutt RL (2003) Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol 26(3):409–420
Storch G, Seiffert C (2007) Extraordinarily preserved specimen of the oldest known glirid from the middle Eocene of Messel (Rodentia). J Vert Palaeontol 27(1):189–194
Acknowledgments
The present work was supported partly by the grant “The Genetic Evaluation of Important Biological Resources (No. 074-1800-1844-304)” funded by the National Institute of Biological Resources, Korean Government, and by the National Research Foundation (NRF) grant “Basic Science Research Program (KRF-2007-313-C00675)” funded by the Ministry of Education, Science and Technology (MEST) awarded to U. W. H. We also thank anonymous reviewers for providing valuable comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ryu, S.H., Kwak, M.J. & Hwang, U.W. Complete mitochondrial genome of the Eurasian flying squirrel Pteromys volans (Sciuromorpha, Sciuridae) and revision of rodent phylogeny. Mol Biol Rep 40, 1917–1926 (2013). https://doi.org/10.1007/s11033-012-2248-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2248-x