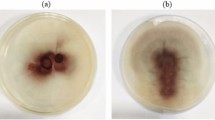
Abstract
Chitinases are glycosyl hydrolases that cleave the β-1,4-glycosidic linkages between N-acetylglucosamine residues in chitin which is a major component of fungal cell wall. Plant chitinases hydrolyze fungal chitin to chitin oligosaccharides that serve as elicitors of plant defense system against fungal pathogens. However, plants synthesize many chitinase isozymes and some of them are not pathogenesis-related. In this study, three full-length cDNA sequences encoding a putative chitinase (EgChit3-1) and two chitinase-like proteins (EgChit1-1 and EgChit5-1) have been cloned from oil palm (Elaeis guineensis) by polymerase chain reaction (PCR). The abundance of these transcripts in the roots and leaves of oil palm seedlings treated with Ganoderma boninense (a fungal pathogen) or Trichoderma harzianum (an avirulent symbiont), and a combination of both fungi at 3, 6 and 12 weeks post infection were profiled by real time quantitative reverse-transcription (qRT)-PCR. Our findings showed that the gene expression of EgChit3-1 increased significantly in the roots of oil palm seedlings treated with either G. boninense or T. harzianum and a combination of both; whereas the gene expression of EgChit1-1 in the treated roots of oil palm seedlings was not significantly higher compared to those of the untreated oil palm roots. The gene expression of EgChit5-1 was only higher in the roots of oil palm seedlings treated with T. harzianum compared to those of the untreated oil palm roots. In addition, the gene expression of EgChit1-1 and EgChit3-1 showed a significantly higher gene expression in the leaf samples of oil palm seedlings treated with either G. boninense or T. harzianum.






Similar content being viewed by others
Abbreviations
- BSR:
-
Basal stem rot
- CBD:
-
Chitin biding domain
- CFU:
-
Colony forming units
- CTAB:
-
Cetyl trimethylammonium bromide
- ESTs:
-
Expressed sequence tags
- G. boninense:
-
Ganoderma boninense
- GH:
-
Glycosyl hydrolase
- MEGA:
-
Molecular evolutionary genetics analysis
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- PR:
-
Pathogenesis-related
- PSA:
-
Potato sucrose agar
- qRT:
-
Quantitative reverse-transcription
- RACE:
-
Rapid amplification of cDNA-ends
- T. harzianum:
-
Trichoderma harzianum
- wpi:
-
Week post infection
References
Kasprzewska A (2003) Plant chitinases–regulation and function. Cell Mol Biol Lett 8:809–824
Boller T, Gehri A, Mauch F, Vögeli U (1983) Chitinase in bean leaves: induction by ethylene, purification, properties and possible function. Planta 157:22–31
Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochem J 293:781–788
Andersen MD, Jensen A, Robertus JD, Skriver K (1997) Heterologous expression and characterization of wild-type and mutant forms of a 26 kDa endochitinase from barley (Hordeum vulgare L.). Biochem J 322:815–822
Funkhouser JD, Aronson NN Jr (2007) Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol Biol 7:96
Neuhaus JM (1999) Chapter 4. Plant chitinases (PR-3, PR-4, PR-8, PR-11). In: Datta SK, Muthukrishnan S (eds) Pathogenesis-related proteins in plants. CRC Press LLC, Boca Raton, pp 77–105
Neuhaus JM, Fritig B, Linthorst HJM, Meins FJ (1996) A revised nomenclature for chitinase genes. Plant Mol Biol Rep 14:102–104
Neuhaus JM, Sticher L, Meins F Jr, Boller T (1991) A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc Natl Acad Sci USA 88:10362–10366
Passarinho PA, de Vries AC (2002) Arabidopsis chitinases: a genomic survey. In: Somerville CR, Meyerowitz EM (eds) The Arabidopsis book. American Society of Plant Biologists, Rockville, pp 1–25
Heitz T, Segond S, Kauffmann S, Geoffroy P, Prasad V, Brunner F, Fritig B, Legrand M (1994) Molecular characterization of a novel tobacco pathogenesis-related (PR) protein: a new plant chitinase/lysozyme. Mol Gen Genet 245:246–254
Ponstein AS, Bres-Vloemans SA, Sela-Buurlage MB, van den Elzen PJ, Melchers LS, Cornelissen BJ (1994) A novel pathogen- and wound-inducible tobacco (Nicotiana tabacum) protein with antifungal activity. Plant Physiol 104:109–118
Legrand M, Kauffmann S, Geoffroy P, Fritig B (1987) Biological function of pathogenesis-related proteins: four tobacco pathogenesis-related proteins are chitinases. Proc Natl Acad Sci USA 84:6750–6754
Kombrink E, Schroder M, Hahlbrock K (1988) Several “pathogenesis-related” proteins in potato are 1,3-β -glucanases and chitinases. Proc Natl Acad Sci USA 85:782–786
Hietala AM, Kvaalen H, Schmidt A, Jøhnk N, Solheim H, Fossdal CG (2004) Temporal and spatial profiles of chitinase expression by norway spruce in response to bark colonization by Heterobasidion annosum. Appl Environ Microbiol 70:3948–3953
Liu JJ, Ekramoddoullah AKM, Zamani A (2005) A class IV chitinase is up-regulated by fungal infection and abiotic stresses and associated with slow-canker-growth resistance to Cronartium ribicola in western white pine (Pinus monticola). Phytopathology 95:284–291
Fossdal CG, Hietala AM, Kvaalen H, Solheim H (2006) Changes in host chitinase isoforms in relation to wounding and colonization by Heterobasidion annosum: early and strong defense response in 33-year-old resistant Norway spruce clone. Tree Physiol 26:169–177
Metraux JP, Burkhart W, Moyer M, Dincher S, Middlesteadt W, Williams S, Payne G, Carnes M, Ryals J (1989) Isolation of a complementary DNA encoding a chitinase with structural homology to a bifunctional lysozyme/chitinase. Proc Natl Acad Sci USA 86:896–900
Mauch F, Staehelin LA (1989) Functional implications of the subcellular localization of ethylene-induced chitinase and β-1,3-glucanase in bean leaves. Plant Cell 1:447–457
Silipo A, Erbs G, Shinya T, Dow JM, Parrilli M, Lanzetta R, Shibuya N, Newman MA, Molinaro A (2010) Glyco-conjugates as elicitors or suppressors of plant innate immunity. Glycobiology 20:406–419
Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K (1993) Plant chitinases. Plant J 3:31–40
Latgé JP (2007) The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol 66:279–290
Santos P, Fortunato A, Ribeiro A, Pawlowski K (2008) Chitinases in root nodules. Plant Biotech 25:299–307
Broglie KE, Gaynor JJ, Broglie RM (1986) Ethylene-regulated gene expression: molecular cloning of the genes encoding an endochitinase from Phaseolus vulgaris. Proc Natl Acad Sci USA 83:6820–6824
Ariffin D, Idris AS, Singh G (2000) Status of Ganoderma in oil palm. In: Flood J, Bridge PD, Holderness M (eds) Ganoderma diseases of perennial crops. CABI Publishing, Wallingford, pp 49–68
Paterson RRM (2007) Ganoderma disease of oil palm—a white rot perspective necessary for integrated control. Crop Prot 26:1369–1376
Nur Ain Izzati MZ, Faridah A (2008) Disease suppression in Ganoderma-infected oil palm seedlings treated with Trichoderma harzianum. Plant Prot Sci 44:101–107
Shafiquzzaman S, Umi KY, Kausar H, Sarwar J (2009) In vitro studies on the potential Trichoderma harzianum for antagonistic properties against Ganoderma boninense. J Food Agric Environ 7:970–976
Ferreira RB, Monteiro S, Freitas R, Santos CN, Chen ZJ, Batista LM, Duarte J, Borges A, Teixeira AR (2007) The role of plant defense proteins in fungal pathogenesis. Mol Plant Pathol 8:677–700
Siswanto SD, Darmono TW (1998) Chitinase and β-1,3- glucanase activities against Ganoderma sp. in oil palm. In: Tahardi JS, Darmono TW, Siswanto SD, Nataatmadja R (eds) Proceedings of the BTIG Workshop on oil palm improvement through biotechnology. Bogor, Indonesia, pp 104–114
van Hengel AJ, Tadesse Z, Immerzeel P, Schols H, van Kammen A, de Vries SC (2001) N-acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol 125:1880–1890
Zhong RQ, Kays SJ, Schroeder BP, Ye ZH (2002) Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14:165–179
Naher L, Ho CL, Tan SG, Umi KY, Faridah A (2011) Cloning of transcripts encoding chitinases from Elaeis guineensis Jacq. and their expression profiles in response to fungal infections. Physiol Mol Plant P 76:96–103
Ho CL, Kwan YY, Choi MC, Tee SS, Ng WH, Lim KA, Lee YP, Ooi SE, Lee WW, Tee JM, Tan SH, Kulaveerasingam H, Sharifah SRSA, Meilina OA (2007) Analysis and functional annotation of expressed sequence tags (ESTs) from multiple tissues of oil palm (Elaeis guineensis Jacq.). BMC Genomics 8:381–392
Wang T, Zhang NH, Du LF (2005) Isolation of RNA of high quality and yield from Ginkgo biloba leaves. Biotechnol Lett 27:629–633
Altschul SF, Madden TL, Schaffer AA, Zhang I, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Tamura K, Dudley J, Nei M, Kumar S (2007) Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Shamala S, Faridah A, Zainal AMA, Umi KY (2008) Efficacy of single and mixed treatments of Trichoderma harzianum as biocontrol agents of Ganoderma basal stem rot in oil palm. J Oil Palm Res 20:470–483
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:1–12
Kuo CJ, Liao YC, Yang JH, Huang LC, Chang CT, Sung HY (2008) Cloning and characterization of an antifungal class III chitinase from suspension-cultured bamboo (Bambusa oldhamii) cells. J Agric Food Chem 56:11507–11514
Ohnuma T, Numata T, Osawa T, Mizuhara M, Lampela O, Juffer AH, Skriver K, Fukamizo T (2011) A class V chitinase from Arabidopsis thaliana: gene responses, enzymatic properties, and crystallographic analysis. Planta 234:123–137
Kim YS, Lee JH, Yoon GM, Cho HS, Park S-W, Suh MC, Choi D, ha JH, Liu JR, Pai HS (2000) CHRK1, a chitinase-related receptor like kinase in tobacco. Plant Physiol 123:905–915
van Damme EJM, Culerrier R, Barre A, Alvarez R, Rougé P, Peumans WJ (2007) A novel family of lectins evolutionarily related to class V chitinases: an example of neofunctionalization in legumes. Plant Physiol 144:662–672
Liu ZH, Yang CP, Qi XT, Xiu LL, Wang YC (2010) Cloning, heterologous expression, and functional characterization of a chitinase gene, Lbchi32, from Limonium bicolor. Biochem Genet 48:669–679
Samac DA, Shah DM (1991) Developmental and pathogen-induced activation of the Arabidopsis acidic chitinase promoter. Plant Cell 3:1063–1072
Lawton KA, Beck J, Potter S, Ward E, Ryals J (1994) Regulation of cucumber class III chitinase gene expression. Mol Plant Microbe Interact 7:48–57
Meier BM, Shaw N, Ślusarenko AJ (1993) Spatial and temporal accumulation of defence gene transcripts in bean (Phaseolus vulgaris) leaves in relation to bacteria-induced hypersensitive cell death. Mol Plant Microbe Interact 6:453–466
Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2:43–56
Ebrahim S, Usha K, Singh B (2011) Pathogenesis related (PR) proteins in plant defense mechanism. In: Méndez-Vilas A (ed) Science against microbial pathogens: communicating current research and technological advances. Formatex, Badajoz, pp 1043–1054
Acknowledgments
We acknowledge the Malaysian Palm Oil Board (MPOB) for funding the research and K. A. Yeoh under the Graduate Studies Assistantship Scheme (GSAS). We thank the Director-General of MPOB for permission to publish this paper, the Pathology Laboratory of MPOB and Mycology Laboratory of UPM for providing us the Ganoderma boninense PER71 and Trichoderma harzianum T32 cultures respectively. We also thank Jugra Palm Oil Mill Sdn. Bhd., Banting for providing us the palm pressed fibres.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yeoh, KA., Othman, A., Meon, S. et al. Sequence analysis and gene expression of putative oil palm chitinase and chitinase-like proteins in response to colonization of Ganoderma boninense and Trichoderma harzianum . Mol Biol Rep 40, 147–158 (2013). https://doi.org/10.1007/s11033-012-2043-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2043-8