Abstract
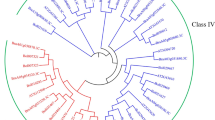
Brassica is a very important vegetable group because of its contribution to human nutrition and consequent economic benefits. However, biotic stress is a major concern for these crops and molecular biology techniques offer the most efficient of approaches to address this concern. Chitinase is an important biotic stress resistance-related gene. We identified three genes designated as Brassica chitinase like protein (BrCLP1), BrCLP2 and BrCLP3 from a full-length cDNA library of Brassica rapa cv. Osome. Sequence analysis of these genes confirmed that BrCLP1 was a class IV chitinase, and BrCLP2 and BrCLP3 were class VII chitinases. Also, these genes showed a high degree of homology with other biotic stress resistance-related plant chitinases. In expression analysis, organ-specific expression of all three genes was high except BrCLP1 in all the organs tested and BrCLP2 showed the highest expression compared to the other genes in flower buds. All these genes also showed expression during all developmental growth stages of Chinese cabbage. In addition, BrCLP1 was up-regulated with certain time of infection by Pectobacterium carotovorum subsp. carotovorum in Chinese cabbage plants during microarray expression analysis. On the other hand, expression of BrCLP2 and BrCLP3 were increased after 6 h post inoculation (hpi) but decreased from 12 hpi. All these data suggest that these three chitinase genes may be involved in plant resistance against biotic stresses.



Similar content being viewed by others
References
Cardoza V, Stewart CN Jr (2004) Brassica biotechnology: progress in cellular and molecular biology. In Vitro Cell Dev Biol Plants 40:542–551
Salunkhe DK, Kadam SS (1998) Handbook of vegetable science technology: production, composition, storage, and processing. Marcel Dekker Inc., New York
Agrios GN (1997) Plant pathology, 4th edn. Academic Press, San Diego
Park YS, Min HJ, Ryang SH, Oh KJ, Cha JS, Kim HY, Cho TJ (2003) Characterization of salicylic acid-induced genes in Chinese cabbage. Plant Cell Rep 21:1027–1034
Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, van den Elzen PJM, Comelissen BJC (1993) Only specific tobacco (Nicotiana tabacum) chitinases and-1, 3-glucanase exhibit antifungal activity. Plant Physiol 101:857–863
Abeles FB, Bossart RP, Forrence LE, Habig WH (1971) Preparation and purification of glucanase and chitinase from bean leaves. Plant Physiol 47:129–134
Muthukrishnan S, Liang GH, Trick HN et al (2001) Pathogenesis-related proteins and their genes in cereals. J Plant Cell Tissue Organ Cult 64:93–114
Wessels JGH, Seitsma JH (1981) Fungal cell walls: a survey. In: Tanner W, Loewus FA (eds) Encyclopedia of plant physiology, new series, vol 133B, plant carbohydrates II. Springer-Verlag, New York, pp 352–394
Roby D, Toppan A, Esquerre-Tugaye MT (1989) Systemic induction of chitinase activity and resistance in melon plants upon fungal infection or elicitor treatment. Physiol Mol Plant Pathol 33:409–417
Wargo PM (1975) Lysis of the cell wall of Armillaria mellea by enzymes from forest trees. Physiol Plant Pathol 5:99–105
Hadwiger LA, Beckman JM (1980) Chitosan as a component of pea-Fusarium solani interactions. Plant Physiol 66:205–211
Cota IE, Troncoso-Rojas R, Sotelo-Mundo R, Sánchez-Estrada A, Tiznado-Hernández ME (2007) Chitinase and b-1, 3-glucanase enzymatic activities in response to infection by Alternaria alternata evaluated in two stages of development in different tomato fruit varieties. Sci Hortic 2591:1–9
Wasano N, Konno K, Nakamura M, Hirayama C, Hattori M et al (2009) A unique latex protein, MLX56, defends mulberry trees from insects. Phytochemistry 70:880–888
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4325
Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Park JI, Kumar TS, Ahmed NU, Nou IS (2010) Construction of full-length cDNA library and investigation of potential genes in Brassica rapa. J Basic Life Res Sci 10:6–11
Guruprasad K, Reddy BV, Pandit MW (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng 4(2):155–161
Neuhaus JM (1999) Plant chitinases (PR-3, PR-4, PR-8, PR-11). In: Datta SK, Muthukrishnan S (eds) Pathogenesis-related protein in plants. CRC Press, Cambridge, pp 77–105
Neuhaus JM, Fritig B, Linthorst HJM, Meins F Jr, Mikkelsen JD, Ryals J (1996) A revised nomenclature for chitinase genes. Plant Mol Bio Rep 14(2):102–104
Henrissat B, Davies G (1997) Structural and sequence-based classification of glycosyl hydrolases. Curr Opin Struct Biol 7:637–644
Cohen-Kupeic R, Chet I (1998) The molecular biology of chitin digestion. Curr Opin Biotechnol 9:270–277
Rasmussen U, Bojsen K, Collinge DB (1992) Cloning and characterization of a pathogen-induced chitinase in Brassica napus. Plant Mol Biol 20(2):277–287
Robinson SP, Jacobs AK, Dry IB (1997) A class IV chitinase is highly expressed in grape berries during ripening. Plant Physiol 114(3):771–778
Troedsson UM, Terefework Z, Jarl-Sunesson CI (2006) Cloning of four chitinase genes and a lectin gene in Galega orientalis. Symbiosis 42(1):25–37
de A Gerhardt LB, Sachetto-Martins G, Contarini MG, Sandroni M, de P Ferreira R, de Lima VM, Cordeiro MC, de Oliveira DE, Margis-Pinheiro M (1997) Arabidopsis thaliana class IV chitinase is early induced during the interaction with Xanthomonas campestris. FEBS Lett 419(1):69–75
Li J, Liu JY (2003) A novel cotton gene encoding a new class of chitinase. Acta Bot Sin 45(12):1489–1496
Kezuka Y, Kitazaki K, Itoh Y, Watanabe J, Takaha O, Watanabe T, Nishizawa Y, Nonaka T (2004) Crystallization and preliminary X-ray analysis of plant class I chitinase from rice. Protein Pept Lett 11(4):401–405
Fan J, Wang H, Feng D, Liu B, Liu H, Wang J (2007) Molecular characterization of plantain class I chitinase gene and its expression in response to infection by Gloeosporium musarum Cke and Massee and other abiotic stimuli. J Biochem 142(5):561–570
Leah R, Tommerup H, Svendsen I, Mundy J (1991) Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem 266(3):1564–1573
Zhang D, Hrmova M, Wan CH, Wu C, Balzen J, Cai W, Wang J, Densmore LD, Fincher GB, Zhang H, Haigler CH (2004) Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol Biol 54(3):353–372
Li DM, Staehelin C, Wang WT, Peng SL (2010) Molecular cloning and characterization of a chitinase-homologous gene from Mikania micrantha infected by Cuscuta campestris. Plant Mol Biol Rep 28(1):90–101
Hamel F, Bellemare G (1995) Characterization of a class I chitinase gene and of wound-inducible, root and flower-specific chitinase expression in Brassica napus. Biochim Biophys Acta 1263(3):212–220
Krishnaveni S, Liang GH, Muthukrishnan S, Manickam A (1999) Purification and partial characterization of chitinases from sorghum seeds. Plant Sci 144(1):1–7
Grison R, Grezes-Besset B, Schneider M, Lucante N, Olsen L, Leguay JJ, Toppan A (1996) Field tolerance to fungal pathogens of Brassica napus constitutively expressing a chimeric chitinase gene. Nat Biotechnol 4:643–646
Oldach KH, Becker D, Lorz H (2001) Heterologous expression of the genes mediating enhanced fungal resistance in transgenic wheat. Mol Plant Microbe Interact 14:832–838
Lawrence SD, Novak NG (2006) Expression of poplar chitinase in tomato leads to inhibition of development in colorado potato beetle. Biotechnol Lett 28:593–599
Kitajima S, Kamei K, Taketani S, Yamaguchi M, Kawai F, Komatsu A, Inukai Y (2010) Two chitinase-like proteins abundantly accumulated in latex of mulberry show insecticidal activity. BMC Biochem 11:6
Libantova J, Kamarainen T, Moravcikova J, Matusikova I, Salaj J (2009) Detection of chitinolytic enzymes with different substrate specificity in tissues of intact sundew (Drosera rotundifolia L.): chitinases in sundew tissues. Mol Biol Rep 36(5):851–856
Neale AD, Wahleithner JA, Lund M, Bonnett HT, Kelly A, Meeks-Wagner DR, Peacock WJ, Dennis ES (1990) Chitinase, [beta]-1, 3-Glucanase osmotin, and extensin are expressed in tobacco explants during flower formation. Plant Cell 2:673–684
Samac DA, Hironaka CM, Yallaly PE, Shah DM (1990) Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol 93:907–914
del Campillo E, Lewis LN (1992) Occurrence of 9.5 cellulase and other hydrolases in flower reproductive organs undergoing major cell wall disruption. Plant Physiol 99:1015–1020
de Jong AJ, Heidstra R, Spanik HP, Hartog MV, Meijer EA, Hendriks T, Loschiavo F, Terzi M, Bisseling T, van Kammen A, de Vries SC (1993) Rhizobium lipooligosaccharides rescue a carrot somatic embryo mutant. Plant Cell 5:615–620
Van Loon LC, Van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
Van Loon LC (2001) The families of pathogenesis-related proteins. In: 6th international work-shop on PR-proteins, Spa, 20–24 May 2001, p 9
Selitrennikoff CP (2001) Antifungal proteins. Appl Environ Microbiol 67:2883–2894
Datta K, Velazhahan R, Oliva N, Ona I, Mew T, Khush GS, Muthukrishnan S, Datta SK (1999) Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor Appl Genet 98:1138–1145
Fagoaga C, Rodrigo I, Conejero V, Hinarejos C, Tuset JJ, Arnau J, Pina JA, Navarro L, Peña L (2001) Increased tolerance to Phytophthora citrophthora in transgenic orange plants constitutively expressing a tomato pathogenesis related protein PR-5. Mol Breed 7:175–185
Bachmann D, Rezzonico E, Retelska D, Chételat A, Schaerer S, Beffa R (1998) Improvement of potato resistance to Phytophthora infestans by overexpressing antifungal hydrolases. In: 5th international workshop on pathogenesis-related proteins. Signalling pathways and biological activities, Aussois, 29 March–2 April 1998
Li WL, Faris JD, Muthukrishnan S, Liu DJ, Chen PD, Gill BS (2000) Isolation and characterization of novel cDNA clones of acidic chitinases and β-1, 3-glucanases from wheat spikes infected by Fusarium graminearum. Theor Appl Genet 102(2–3):353–362
Acknowledgments
This study was supported by the Cabbage Genomics assisted breeding supporting center research program funded by Ministry for Food, Agriculture, Forestry and Fisheries of the Korean Government. We wish to thank Genomics Division, Department of Agricultural Bio-resources, National Academy of Agricultural Science, Republic of Korea for providing microarray data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Nasar Uddin Ahmed and Jong-In Park contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ahmed, N.U., Park, JI., Seo, MS. et al. Identification and expression analysis of chitinase genes related to biotic stress resistance in Brassica . Mol Biol Rep 39, 3649–3657 (2012). https://doi.org/10.1007/s11033-011-1139-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1139-x