Abstract
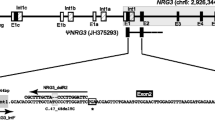
Lmbr1 is the key candidate gene for limb development. Until now, at least five and four alternative splicing isoforms of Lmbr1 gene have been found in human and mouse, respectively. However, only two alternative splicing isoforms of this homologous gene have been reported in chicken. In the present study, one novel chicken Lmbr1 transcript variant (designated Lmbr1-1) was identified by 5′ RACE and RT-PCR. Chicken Lmbr1-1 possesses one novel transcription start site different from Lmbr1-N, and was predicted to encode one 192 amino acid protein with length variation in comparison with chicken LMBR1-N protein, which was produced by 5′ spliced site variation of chicken Lmbr1-N exon 10. Comparing with Lmbr1-N transcript, chicken Lmbr1-1 exhibited restricted tissue distribution of the expression. Comparative sequence analysis revealed a highly conservative intron element between chicken and mammalians from the intron 9 of chicken Lmbr1-N, indicating their possible importance as intronic elements in the regulation of alternative splicing of Lmbr1 in vertebrates. By direct PCR sequencing the exon 10 and its flanking sequences in chicken Lmbr1-N, four variation sites/haplotypes were identified from six chicken breeds. One 797A/G nonsynonymous mutation (266Arg/Gln) locating in exon 10 of chicken Lmbr1-N was predicted to affect the exon splice enhancer motif for serine/arginine-rich protein recognition. These data demonstrated that chicken Lmbr1 was alternatively spliced to generate multiple splice forms, as was the case in mammals and each of the alternative splicing isoforms might function differentially.




Similar content being viewed by others
References
Heus HC, Hing A, van Baren MJ et al (1999) A physical and transcriptional map of the preaxial polydactyly locus on chromosome 7q36. Genomics 57:342–351. doi:10.1006/geno.1999.5796
Clark RM, Marker PC, Kingsley DM (2000) A novel candidate gene for mouse and human preaxial polydactyly with altered expression in limbs of Hemimelic extra-toes mutant mice. Genomics 67:19–27. doi:10.1006/geno.2000.6225
Balci S, Demirtas M, Civelek B, Piskin M, Sensoz O, Akarsu AN (1999) Phenotypic variability of triphalangeal thumb-polysyndactyly syndrome linked to chromosome 7q36. Am J Med Genet 87:399–406. doi:10.1002/(SICI)1096-8628(19991222)87:5<399:AID-AJMG6>3.0.CO;2-A
Dobbs MB, Dietz FR, Gurnett CA, Morcuende JA, Steyers CM, Murray JC (2000) Localization of dominantly inherited isolated triphalangeal thumb to chromosomal region 7q36. J Orthop Res 18:340–344. doi:10.1002/jor.1100180303
Heutink P, Zguricas J, van Oosterhout L et al (1994) The gene for triphalangeal thumb maps to the subtelomeric region of chromosome 7q. Nat Genet 6:287–292. doi:10.1038/ng0394-287
Sato D, Liang D, Wu L et al (2007) A syndactyly type IV locus maps to 7q36. J Hum Genet 52:561–564. doi:10.1007/s10038-007-0150-5
Zguricas J, Heus H, Morales-Peralta E et al (1999) Clinical and genetic studies on 12 preaxial polydactyly families and refinement of the localisation of the gene responsible to a 1.9 cM region on chromosome 7q36. J Med Genet 36:32–40. doi:10.1136/jmg.36.1.33
Escamilla MA, DeMille MC, Benavides E et al (2000) A minimalist approach to gene mapping: locating the gene for acheiropodia, by homozygosity analysis. Am J Hum Genet 66:1995–2000. doi:10.1086/302921
Ianakiev P, van Baren MJ, Daly MJ et al (2001) Acheiropodia is caused by a genomic deletion in C7orf2, the human orthologue of the Lmbr1 gene. Am J Hum Genet 68:38–45. doi:10.1086/316955
Huang Y, Du X, Deng X et al (2007) Single nucleotide polymorphisms in chicken lmbr1 gene were associated with chicken growth and carcass traits. Sci China C Life Sci 50:62–69. doi:10.1007/s11427-007-2033-6
Huang YQ, Deng XM, Du ZQ et al (2006) Single nucleotide polymorphisms in the chicken Lmbr1 gene are associated with chicken polydactyly. Gene 374:10–18. doi:10.1016/j.gene.2005.07.047
Lettice LA, Heaney SJ, Purdie LA et al (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12:1725–1735. doi:10.1093/hmg/ddg180
Lettice LA, Horikoshi T, Heaney SJ et al (2002) Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA 99:7548–7553. doi:10.1073/pnas.112212199
Chang DT, Lopez A, von Kessler DP et al (1994) Products, genetic linkage and limb patterning activity of a murine hedgehog gene. Development 120:3339–3353
Wang ET, Sandberg R, Luo S et al (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476. doi:10.1038/nature07509
Clark RM, Marker PC, Roessler E et al (2001) Reciprocal mouse and human limb phenotypes caused by gain- and loss-of-function mutations affecting Lmbr1. Genetics 159:715–726
Stephens M, Donnelly P (2003) A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169. doi:10.1086/379378
Mignone F, Grillo G, Licciulli F et al (2005) UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 33:D141–D146. doi:10.1093/nar/gki021
Pesole G, Grillo G, Larizza A, Liuni S (2000) The untranslated regions of eukaryotic mRNAs: structure, function, evolution and bioinformatic tools for their analysis. Brief Bioinf 1:236–249. doi:10.1093/bib/1.3.236
Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S (2001) Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276:73–81. doi:10.1016/S0378-1119(01)00674-6
Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM (2006) Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J 25:2792–2801. doi:10.1038/sj.emboj.7601159
Duchow HK, Brechbiel JL, Chatterjee S, Gavis ER (2005) The nanos translational control element represses translation in somatic cells by a Bearded box-like motif. Dev Biol 282:207–217. doi:10.1016/j.ydbio.2005.03.025
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31:3568–3571. doi:10.1093/nar/gkg616
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415. doi:10.1038/ng.259
Horikoshi T, Endo N, Shibata M, Heutink P, Hill RE, Noji S (2003) Disruption of the C7orf2/Lmbr1 genic region is associated with preaxial polydactyly in humans and mice. J Bone Miner Metab 21:1–4. doi:10.1007/s007740300000
Maas SA, Fallon JF (2004) Isolation of the chicken Lmbr1 coding sequence and characterization of its role during chick limb development. Dev Dyn 229:520–528. doi:10.1002/dvdy.10502
Hare MP, Palumbi SR (2003) High intron sequence conservation across three mammalian orders suggests functional constraints. Mol Biol Evol 20:969–978. doi:10.1093/molbev/msg111
Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298. doi:10.1038/nrg775
Blencowe BJ (2000) Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci 25:106–110. doi:10.1016/S0968-0004(00)01549-8
Yan XB, Tang CH, Huang Y et al (2010) Alternative splicing in exon 9 of glucocorticoid receptor pre-mRNA is regulated by SRp40. Mol Biol Rep 37:1427–1433. doi:10.1007/s11033-009-9529-z
Acknowledgments
Financial support for this research was provided by National Natural Science Foundation of China (30771533), the State Major Basic Research Development Program (G20000161) and Innovation Scientists and Technicians Troop Construction Projects of Zhengzhou City (096SYJH16092).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, Y., Chen, W., Li, N. et al. Identification of an alternative splicing isoform of chicken Lmbr1 . Mol Biol Rep 38, 4397–4403 (2011). https://doi.org/10.1007/s11033-010-0567-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0567-3